Multiplex PCR detection primers and multiplex PCR detection method for Escherichia coli O<157>
A technology for the detection of Escherichia coli, which is applied in the field of microbial molecular biology detection, can solve the problems of poor specificity, low sensitivity, and long identification time, and achieve the effect of improving sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
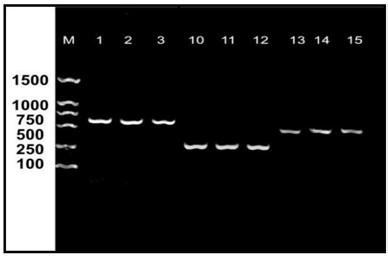
[0029] Example 1: O 157 Design and synthesis of PCR detection primers
[0030] Primers were found in Genbank for Escherichia coli O 157 The sequences of the stx1, flic, rfbE genes of the target gene were compared and analyzed with CLUSTAL, DNAMAN and other biological software to find the conserved region sequence of each target gene, and the conserved region sequence was used as the PCR amplification region, using Primers Premier, The primers were designed by biological software such as Oligo, the specificity and feasibility of the primers were verified by online BLAST, and the primers were synthesized according to conventional methods in the field.
[0031] The primer sequences are:
[0032] rfbE upstream primer F1: 5'-CGTAAGAGGGACCGTAGA-3' (SEQ ID No.1),
[0033] rfbE downstream primer F2: 5'-TGGCTGGATTATCGTTTG-3' (SEQ ID No.2),
[0034] fliC upstream primer F3: 5'-GCTGTCGAGTTCTATCGAG-3' (SEQ ID No.3),
[0035] FliC downstream primer F4: 5'-GTGACTTTATCGCCATTCC-3' (SEQ I...
Embodiment 2
[0039] Example 2: O 157 PCR detection annealing temperature optimization
[0040] 1. Escherichia coli O 157 DNA Extraction
[0041] Escherichia coli O was lysed by boiling 157 (ATCC35150), using phenol-chloroform-isoamyl alcohol reagent to extract bacterial DNA. The specific steps are: take O 157 Put 1ml of bacterial solution in a 1.5ml centrifuge tube and boil in a water bath for 10 minutes. Centrifuge at 10000r / min for 5min, take the supernatant, add an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1) respectively, then mix slightly, leave at room temperature for 5-10min, then centrifuge at 12000rpm for 10min. Repeat the extraction 2-3 times. Aspirate the supernatant, add an equal volume of absolute ethanol, place it at low temperature for 30 minutes, and then centrifuge at 12000 r / min for 10 minutes to retain the precipitate. Wash the DNA with pre-cooled 70% ethanol to remove excess impurities. Centrifuge at 12000r / min for 10min, pour off the upper laye...
Embodiment 3
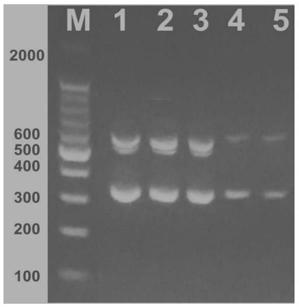
[0050] Example 3: Effect of Primer Ratio on Multiplex PCR Detection
[0051] E. coli O 157 The DNA extraction, PCR reaction system and amplification procedures refer to Example 2. In the PCR reaction system, only the ratio of the primer pair is changed. The molar ratio of the upstream and downstream primers in each pair of primers is 1:1, and the molar ratio between the primer pairs of stx1, flic, and rfbE is respectively set to 1:1: 1. 1:2:1, 2:3:1, research primers for Escherichia coli O 157 The effect of multiplex PCR detection.
[0052] The experimental results found that the brightness of the three bands was better when the molar ratio between the primer pairs of stx1, flic, and rfbE was 1:2:1 and 2:3:1 than when the molar ratio was 1:1:1, indicating that this The three pairs of primers have different amplification efficiencies for their respective target genes.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com