Biomarker and method for predicating relapse and mortality risk of renal cell carcinoma
A biomarker, renal cell carcinoma technology, applied in the direction of biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems that have not been reported, and achieve the effect of good management, efficient recurrence and death risk
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] 1 Implementation samples and methods
[0055] 1.1 Case samples
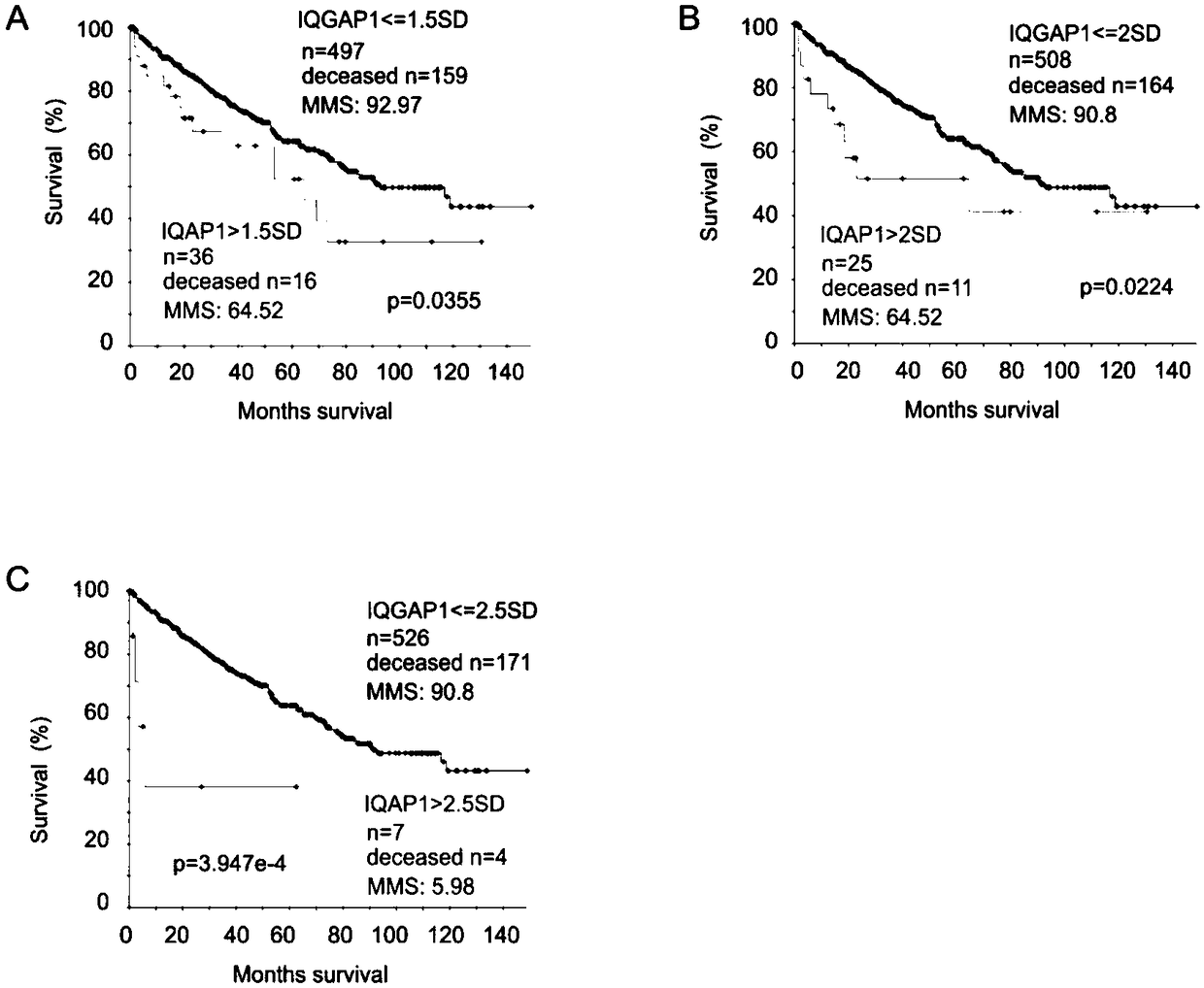
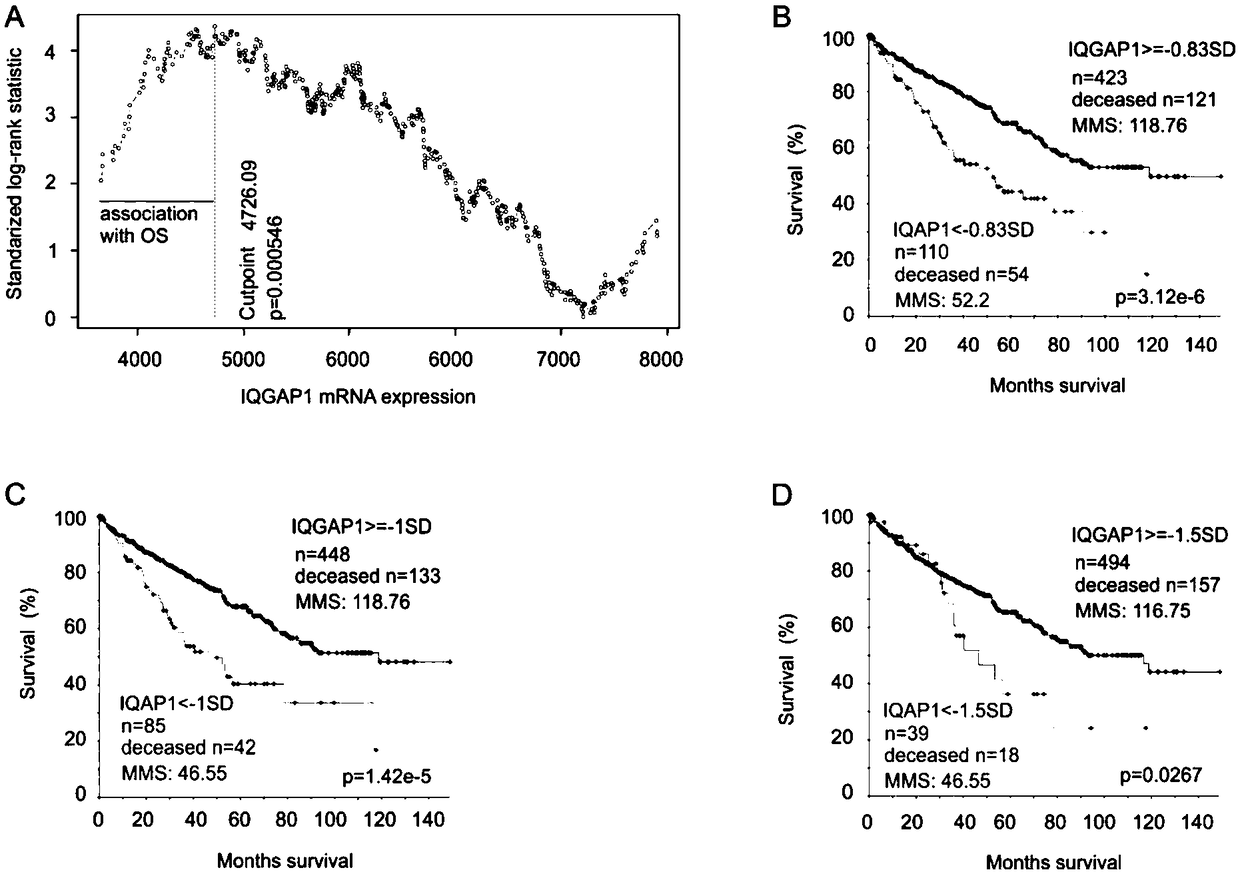
[0056] Select 533 patients with carcinoma in situ from the TCGA Provisional ccRCC database in the cBioPortal database. All tumors were surgically resected and the RNA expression was obtained by RNA sequencing, as well as 140-month follow-up information and some clinical data. 291 tumors / patients in the TCGA Provisional pRCC database were selected for RNA sequencing, as well as follow-up information and some clinical data. The demographic data of ccRCC patients in TCGA are shown in Table 1, where Q is quartile, NA is not available data (not available), and Tart Mol therapy is targeted molecular therapy:
[0057] Table 1
[0058]
[0059]
[0060] 1.2 Gene enrichment analysis
[0061] The enriched gene sets and pathways were analyzed using the GAGE(49) and Reactome(50) language packages in the R language. The database uses KEGG (Kyoto Encyclopedia of Genes and Genomes) and GO (gene ontology) databa...
Embodiment 2
[0090] Used implementation sample, method are identical with embodiment 1;
[0091] Using Sig0.83sub1 and Sig0.83sub2 to predict cRCC mortality risk:
[0092] The HR of SPACA6, TROAP, CENPW and NCOA4 in Sig0.83 is higher, and it is separated as the first subgenome Sig0.83sub1 of Sig0.83, and the DEGs with P value>2.79e-9 are excluded from the remaining genes Elimination, leaving 21 genes to form Sig0.83sub2;
[0093] The sensitivity / specificity / PPV / MMS / P values of Sig0.83sub1, Sig0.83sub2 and Sig0.83cut in predicting the risk of death in ccRCC were 41.1% / 93.6% / 75.8% / 27.2 / 0 (eg Figure 6 D), 62.3% / 83.8% / 65.3% / 36.5 / 0 (such as Figure 6 E) and 50.9% / 90.2% / 71.8% / 31.1 / 0 (as Figure 6 C shown). The tAUCs of Sig0.83sub1 and Sig0.83sub2 for identifying ccRCC mortality risk were greater than 70% and 76% in the four time frames, respectively (eg Figure 6 shown in A).
Embodiment 3
[0095] Used implementation sample, method are identical with embodiment 1;
[0096] Using Sigcmbn to predict ccRCC mortality risk:
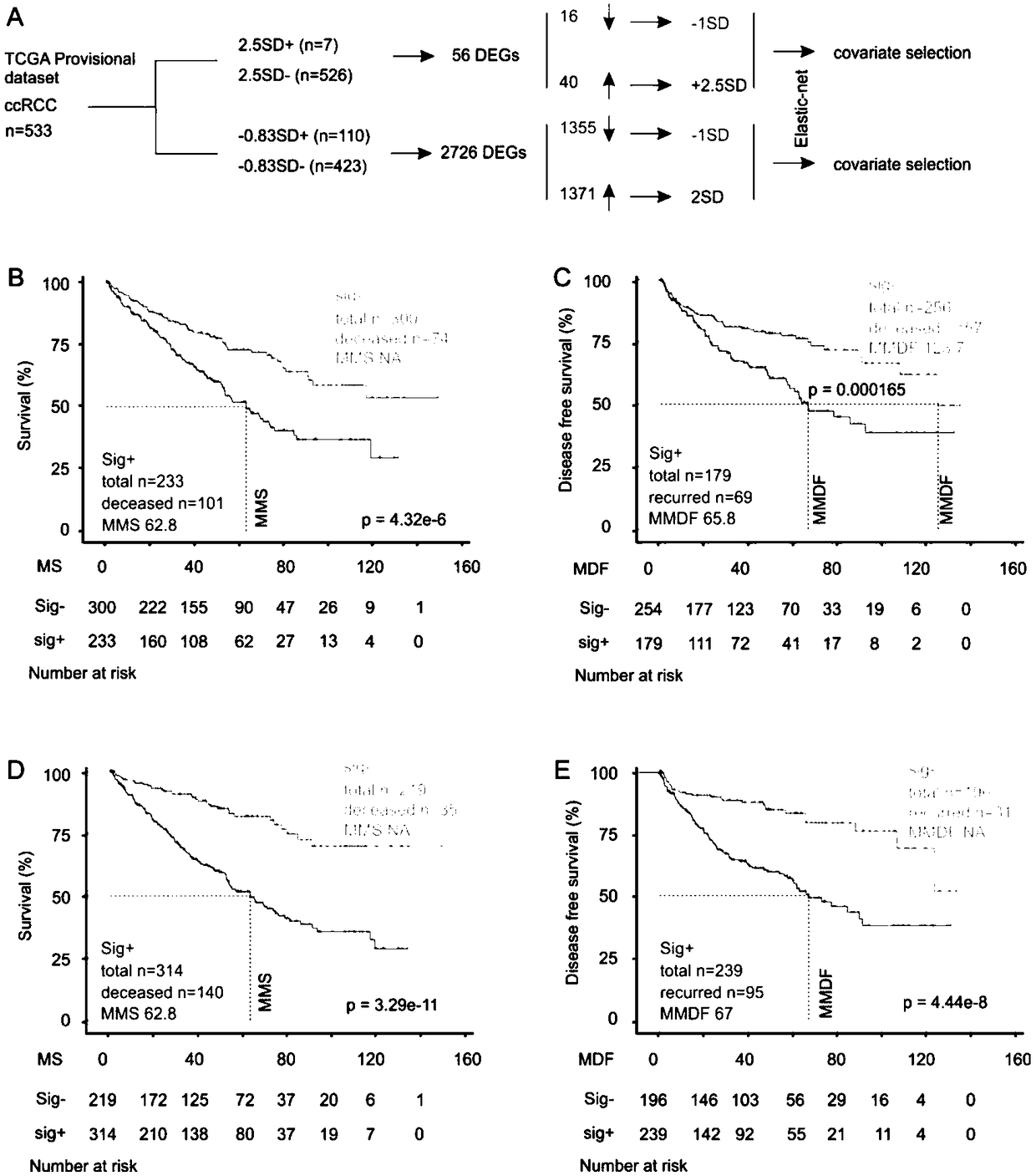
[0097] Sigcmbn predicts tAUC of ccRCC high mortality risk as 78.1% / 13.8M, 78.7% / 31M, 77.8% / 48.9M and 81.3% / 70.6M, higher than Sig2.5 or Sig0.83 (eg Figure 11 A and Figure 6 Shown in A); the cut point score of Sigcmbn (such as Figure 11 B), Q1, median, mean and Q3 (as shown in Figure 12 shown) are all related to the decline in overall survival rate. If Q1, median, Q3 and cut point scores are used to predict the risk of death together, the sensitivity / specificity / PPV / MMS / P value can reach the best 90.3% / 89.1 % / 71.2% / 31.1 / 0 (eg Figure 11 C shown);
[0098] Among the 37 constituent genes of Sigcmbn, the cut point scores of 25 genes are all associated with the decrease of overall survival rate (p≤0.092), and the tAUC of the cumulative score predicting the risk of death is 76% / 23.2M, 82.1% / 36.3M , 86.5% / 45.5M and 80.7% / 58.9M (eg Figure 13 As...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com