Detection method of oxidation modified nitrogenous bases
A detection method, nitrogen base technology, applied in the field of DNA modification detection, can solve the problem of indetermination of 8-oxoG modification damage, etc., and achieve the effect of convenient detection and precise treatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] The method for positioning and detecting base (G) oxidation modification in a DNA sequence based on SMRT sequencing technology, specifically comprises the following steps:
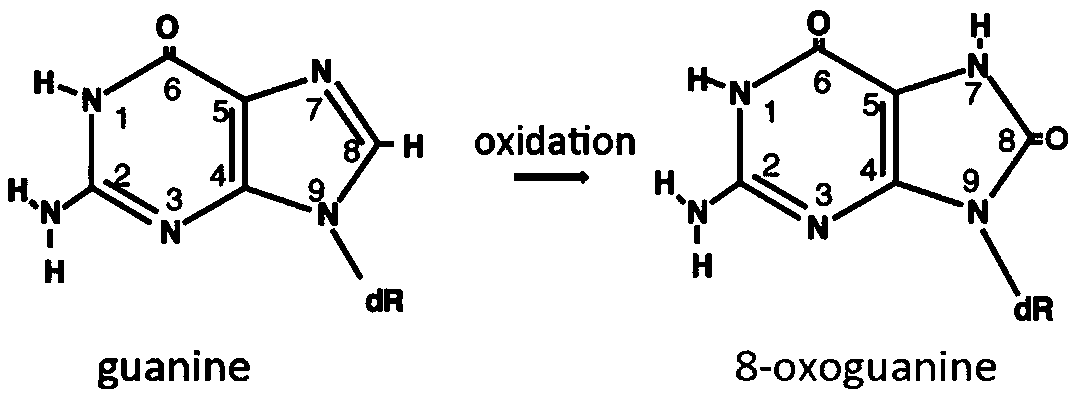
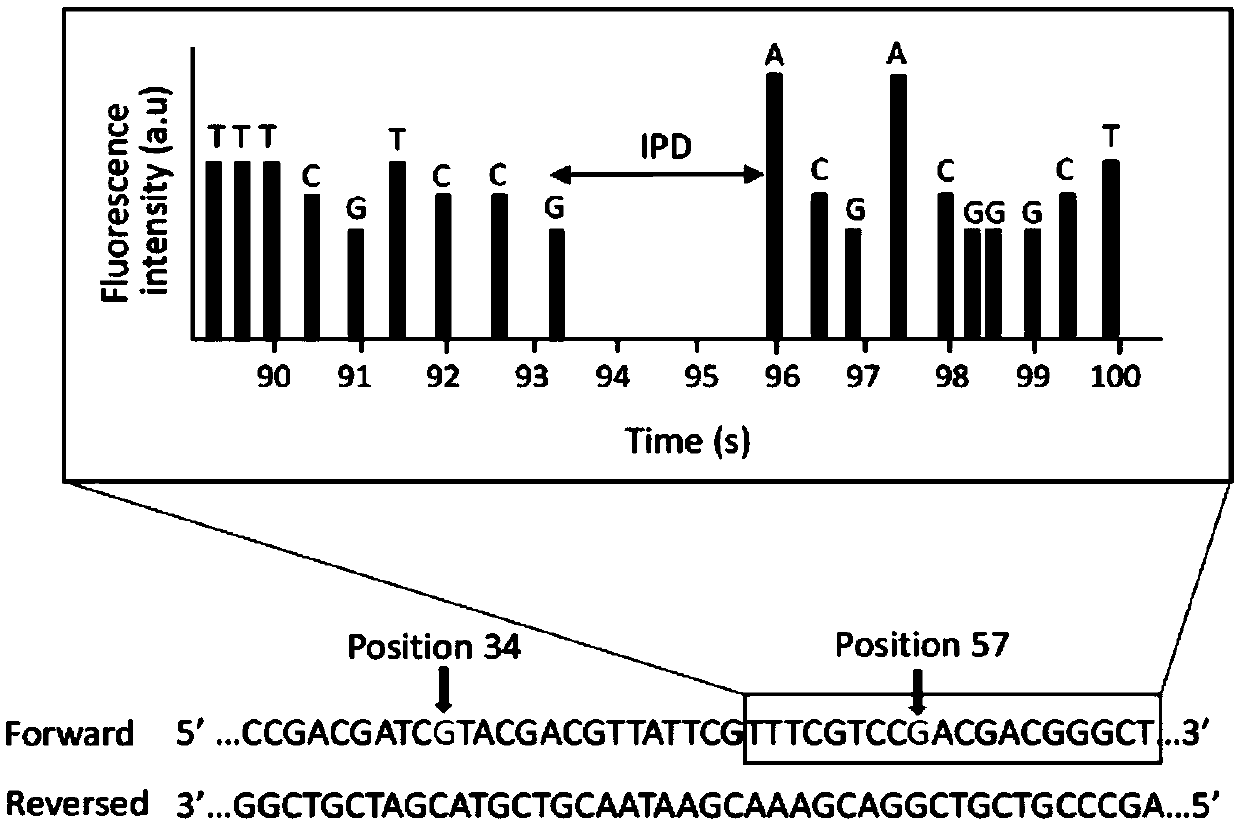
[0041] 1) Using a 155bp long DNA sequence produced by Takara Clontech Company with two 8-oxoG sites, two oxidatively modified guanines are located at the 34th and 57th positions in the upstream of the DNA sequence, and the DNA oxidation modification product 8-hydroxyl The chemical structure of deoxyguanine is as figure 1 shown;
[0042] 2) The sequence sample obtained in step 1) is constructed into a gene library;
[0043] 3) The library of step 2) is sequenced on the PacBio RSII sequencer to obtain the original sequence;
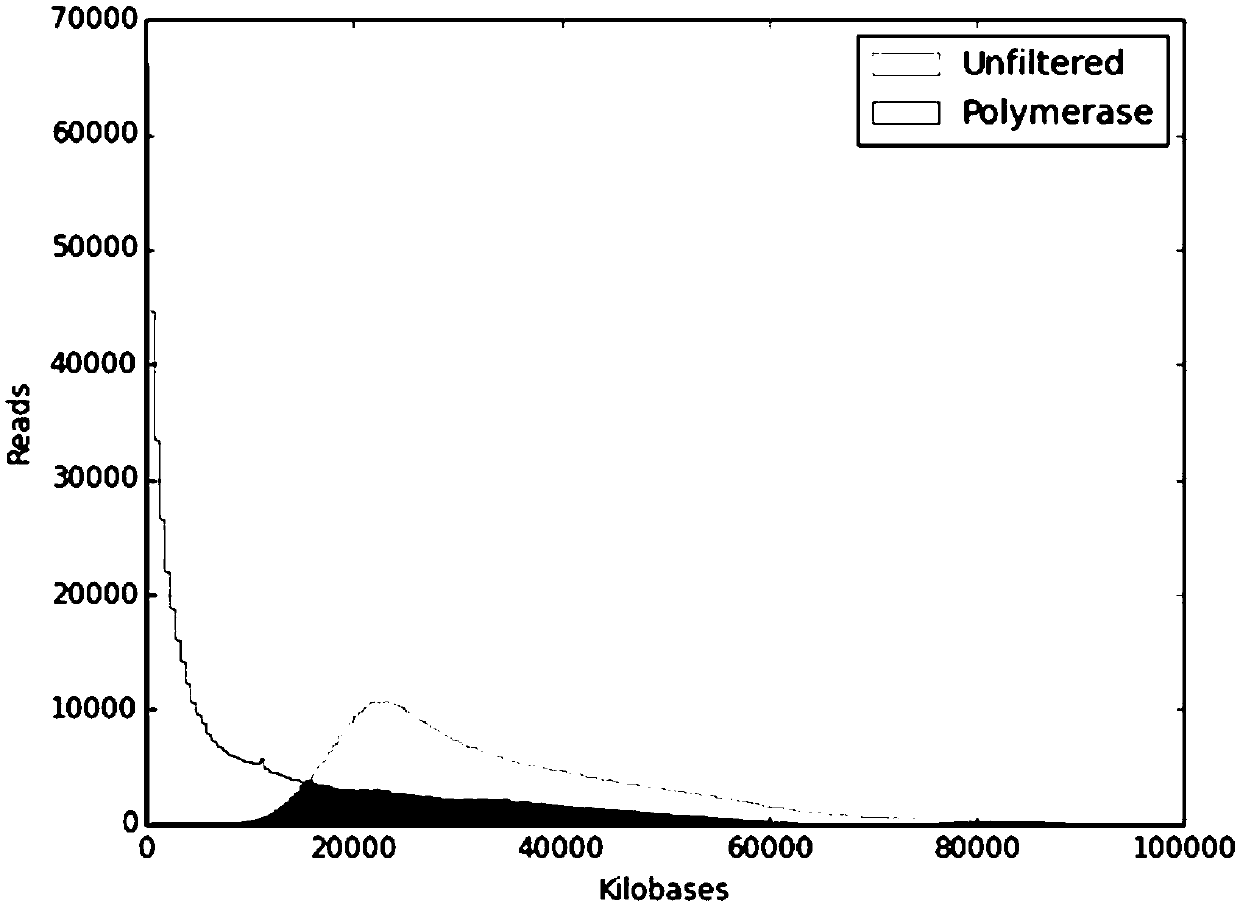
[0044]4) Step 3) The original sequence uses SMRT Link v5.1.0 software for data quality control and filtering, and then the sequence is exported for bioinformatics analysis;
[0045] 5) Use a known synthetic sequence as a reference sequence, step 4) sequence and reference sequ...
Embodiment 2
[0052] The method for identifying the oxidative modification site of plasmid DNA pcDNA3.1 8-oxoG by using SMRT sequencing technology and base modification site analysis method specifically includes the following steps:
[0053] 1) Extract plasmid pcDNA3.1 (5477bp), using Tiangen High Purity Plasmid Extraction Kit (non-centrifugal column type, DP116). The specific method is as follows:
[0054] a) Take 100 mL of overnight cultured bacterial solution, and centrifuge at room temperature at 10,000 rpm for 3 minutes to collect the bacteria.
[0055] b) Aspirate the supernatant as much as possible, add 10mL solution P1 to the centrifuge tube with the cell pellet left, and vortex to thoroughly suspend the cell pellet.
[0056] c) Add 10 mL of solution P2 to the centrifuge tube, immediately turn it up and down gently 6-8 times to fully lyse the bacteria, and place it at room temperature for 5 minutes.
[0057] d) Add 10mL solution P4 to the centrifuge tube, immediately turn it gentl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com