Compositions and methods for the rapid differential detection of zika virus
A technology for Zika virus and virus genome, which is applied in the field of compositions and methods for rapid differential detection of Zika virus, and can solve problems such as methods or assays without differential detection of arboviruses, difficulties in differential diagnosis and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
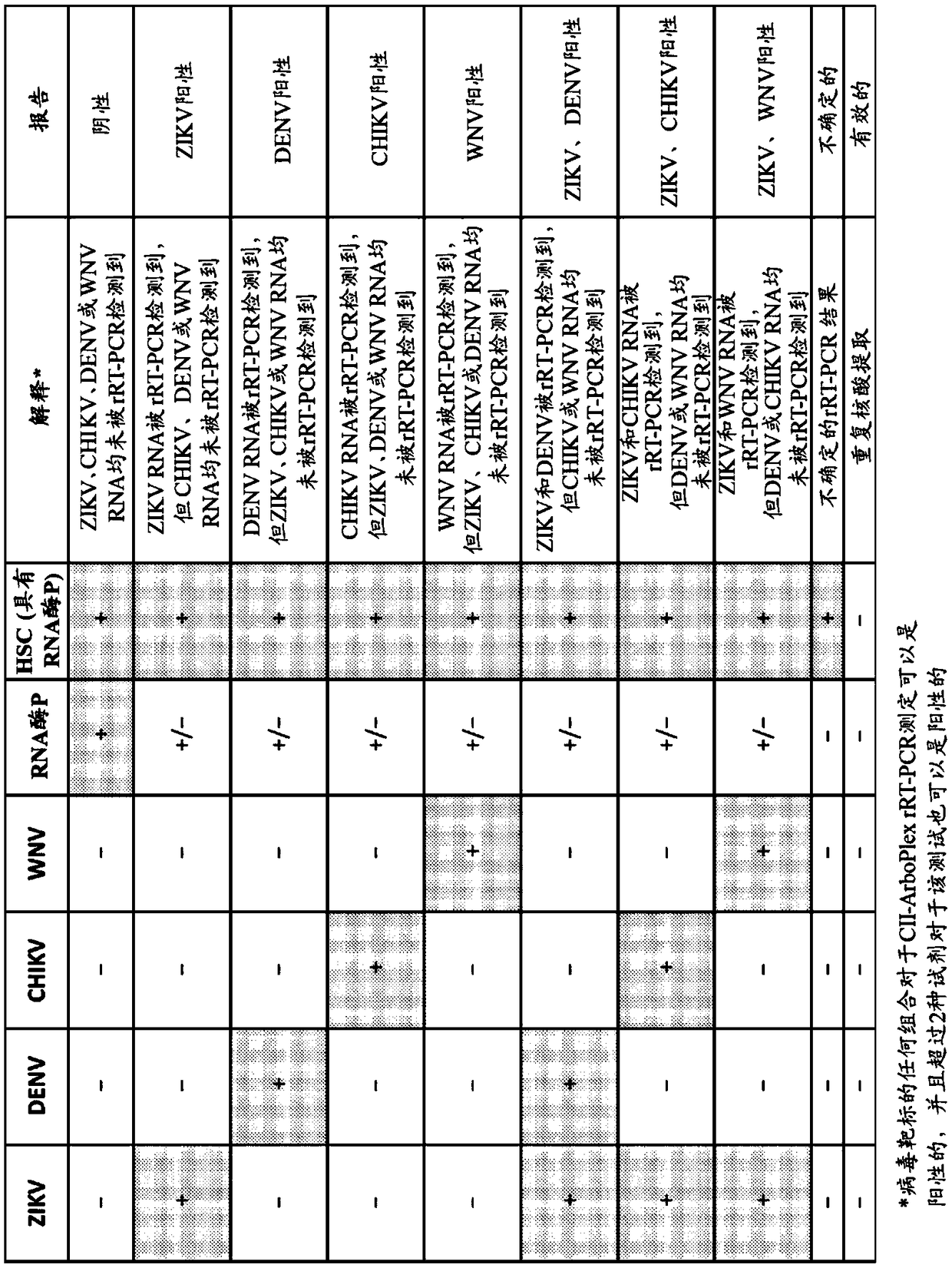
[0137] Example 1 - Primers and Probes for Assays
[0138] The following probes and primers were used in the examples.
[0139] Table 2 - Primers and Probes
[0140]
[0141]
[0142] All primers and probes are generic. Zika primers and probes target the 3'UTR of ZIKV genomic RNA (SEQ ID NO: 1-3). West Nile virus primers and probes target the NS5 gene of WNV genomic RNA (SEQ ID NO: 4-6). Dengue virus primers and probes target the 3'UTR of DENV genomic RNA (SEQ ID NO: 7-10). The gene target of the Chikungunya virus primers and probes was the NSP2 gene (SEQ ID NO: 11-13) of the CHIKV genomic RNA.
[0143] SEQ ID NO: 14-16 are control primers and probes. The gene target is human ribonuclease P subunit p30 mRNA.
Embodiment 2
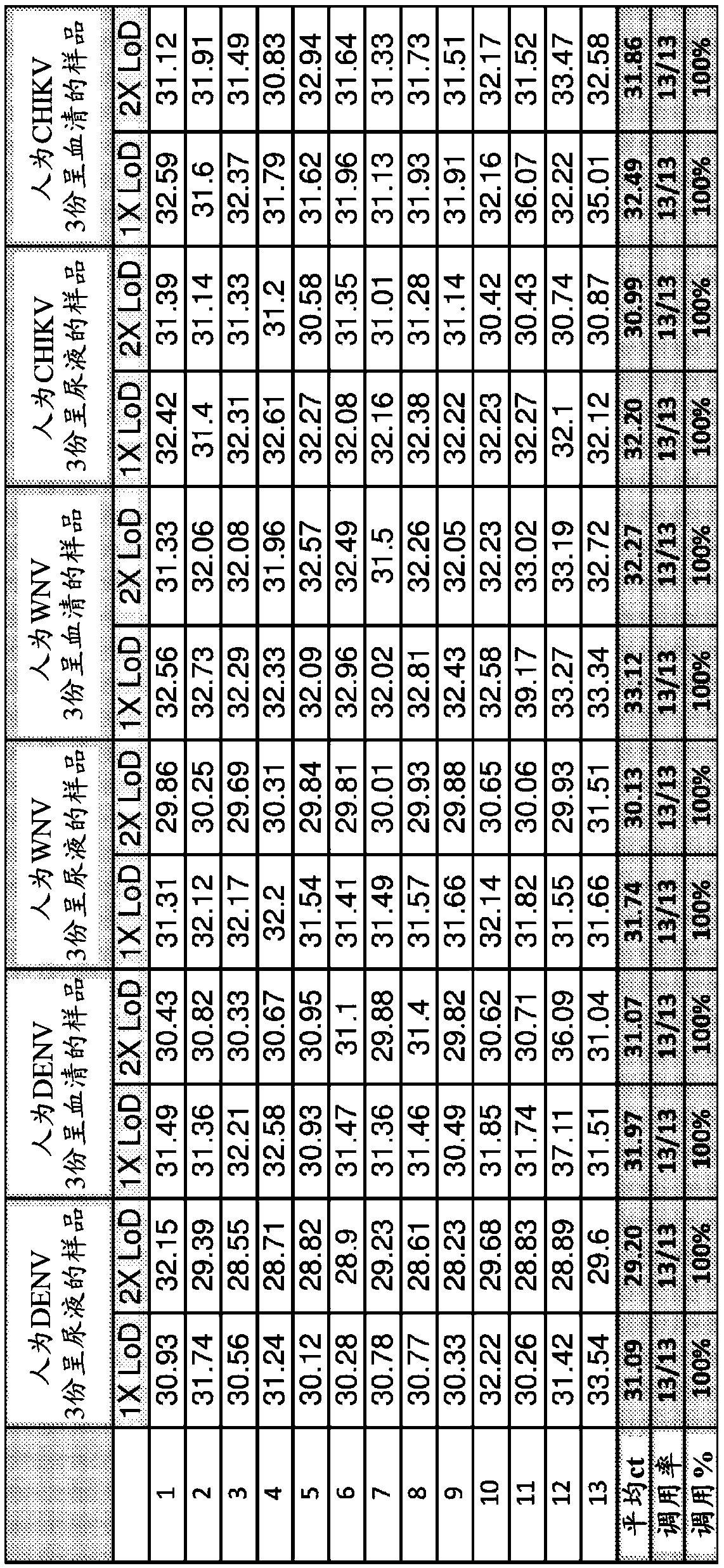
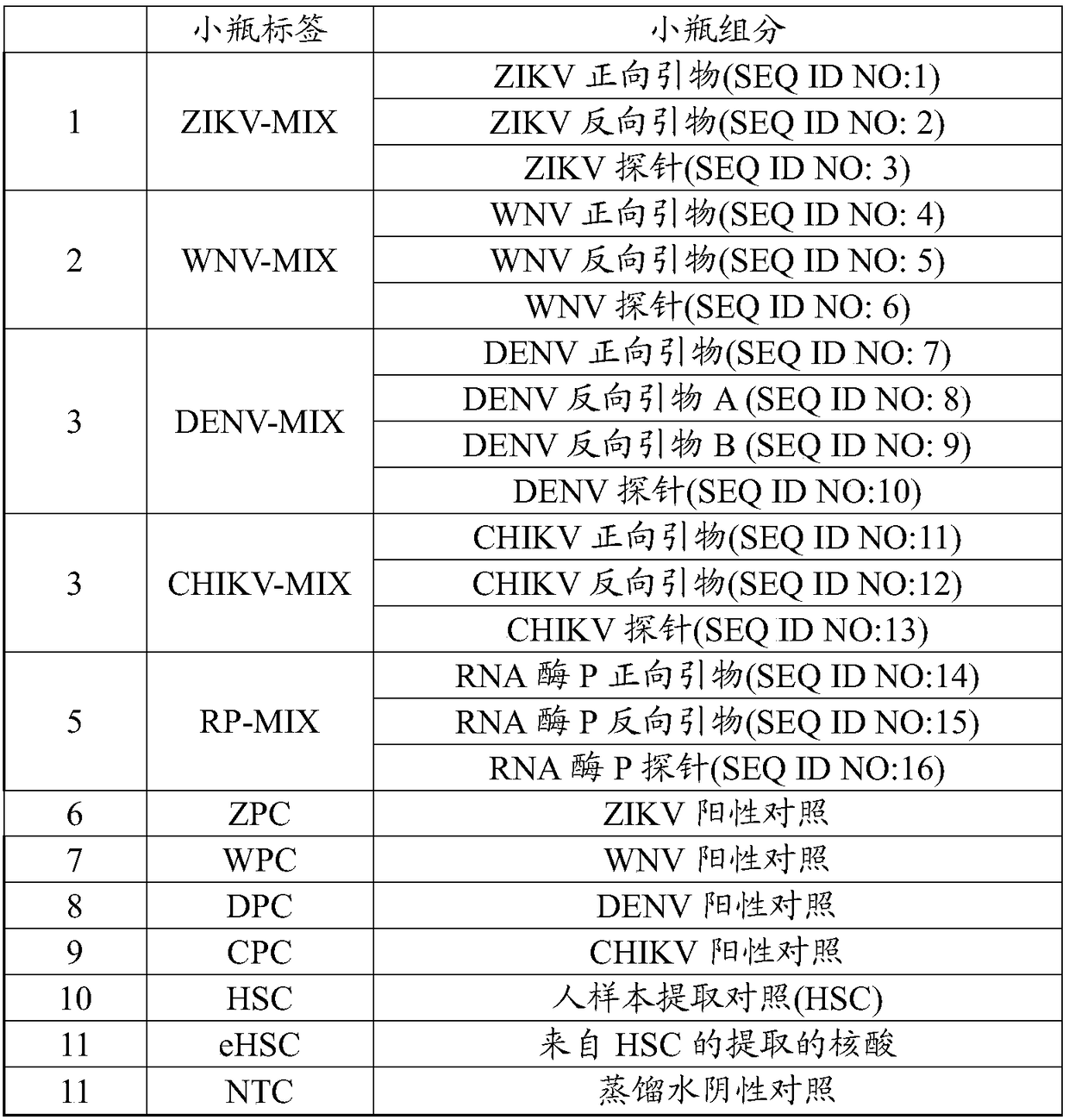
[0144] Example 2 - Reagents and materials for multiplex assays
[0145] Probes and primers listed in Table 2 are supplied in lyophilized form in one vial of each virus to be prepared in 100 μl H 2 Recovery in O. Vials labeled ZIKV-MIX contained primers and probes SEQ ID NO: 1-3. Vials labeled WNV-MIX contained primers and probes SEQ ID NO: 4-6. Vials labeled DENV-MIX contained primers and probes SEQ ID NO: 7-10. Vials labeled CHIK-MIX contained primers and probes of SEQ ID NO: 11-13. Vials labeled RP-MIX contained primers and probes of SEQ ID NO: 14-16. See Tables 1 and 2.
[0146] Additional reagents included assay controls in labeled vials, as shown in Tables 1 and 3.
[0147]All assay controls should be included on each plate and tested concurrently with the clinical samples. These assay controls include extraction control reagents, positive controls for each virus, etc.
[0148] The extraction control was a human sample control (HSC, extraction control) and was a...
Embodiment 3
[0162] Example 3 - Nucleic acid extraction and storage
[0163] manufacturer to The instructions generally follow the extraction process and machine interface operation.
[0164] Briefly, a sample input volume of 250 μL and an elution volume of 50 μL total nucleic acid were used.
[0165] Thaw previously collected and frozen biological samples (including urine and serum) on ice. Pipette 250 μL sample and mix with 750 μL Lysis buffer was mixed and incubated at room temperature for 10-15 min (standard ratio 1:3 sample:lysis buffer).
[0166] 50μL Magnetic silica beads were added to the sample + lysis buffer mixture and mixed well by pipetting sample and magnetic silica beads up and down. Transfer samples to in the sample box. The magnetic silica cartridge is loaded into the machine along with the pipette cartridge and the sample is processed.
[0167] When the run is complete, transfer 50 µL of the eluted total nucleic acid to a clean reaction tube for the PCR assa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com