Prostate cancer early diagnosis or prognosis evaluation marker lncRNA malat1 and application thereof
A technology for prostate cancer and prognosis evaluation, applied in the field of medical diagnosis, can solve the problems of early diagnosis of unexposed prostate cancer, undisclosed relationship between prostate cancer, etc., and achieves the effect of high sensitivity, specificity, and good indication effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] The main purpose of this example is to detect the content of each malat1 fragment of exosome-derived malat1 in serum, specifically including:
[0043] 1. Use an exosome extraction kit (QIAGEN: exoEasy Maxi Kit or miRCURY ExosomeKits; Thermofisher: Total Exosome Isolation Reagent from serum) to extract the total exosomes in the serum. The extracted exosomes were extracted with a ribonucleic acid extraction kit (QIAGEN company: miRNeasy Micro Kit) to extract all the ribonucleic acids in the exosomes. The extracted ribonucleic acid was converted into cDNA using a reverse transcription kit (Takara: PrimeScript™ II 1st Strand cDNA Synthesis Kit).
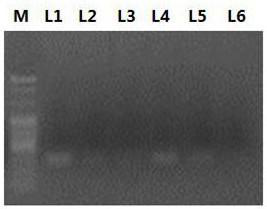
[0044] 2. Design primers for amplifying different fragments on malat1 according to the nucleotide sequence of malat1. The specific primer information is shown in Table 1. Utilize the primers shown in table 1 and carry out PCR reaction and agarose gel electrophoresis to described cDNA (electrophoresis result sees figure 1 ). fig...
Embodiment 2
[0048] The main purpose of this embodiment is to establish a method for quantitative detection of exosome malat1-1 fragments, the method comprising:
[0049] 1. Extract serum total exosomes and ribonucleic acid, and reverse transcribe the ribonucleic acid into cDNA (see Example 2 for specific methods).
[0050] 2. According to the malat1-1 sequence design, the probe used in conjunction with the upstream and downstream primers of malat1-1: wherein, the nucleotide sequence of the malat1-1 is:
[0051] TAGGGGATTTCAGGATTGAGAAATTTTTCCATCGAGCCTTTTTAAAATTGTAGGACTTGT (SEQ ID NO: 13)
[0052] The nucleotide sequence of the probe is: TCCATCGAGCCTTTT (SEQ ID NO: 14), the 5' end of the probe is marked with a FAM fluorescent group, and the 3' end is marked with an MGB modification group.
[0053] 3. A nucleic acid fragment of artificially synthesized malat1 is used as a nucleic acid standard, and diluted to a concentration of 5pg / μL, 1pg / μL, 0.5pg / μL, 0.2pg / μL, 0.1pg / μL, and the nucleic a...
Embodiment 3
[0057] The main purpose of this example is to evaluate the diagnostic effect of serum exosome-derived malat1-1 on early prostate cancer, mainly including:
[0058] 1. Call 299 subjects whose clinical diagnosis results have been confirmed, see Table 2 for specific information.
[0059] 2. Detect the serum exosome-derived malat1 of 299 subjects (see Example 2 for the detection method) and plasma MD-miniRNA (see the Chinese invention patent CN103146688B for the detection method).
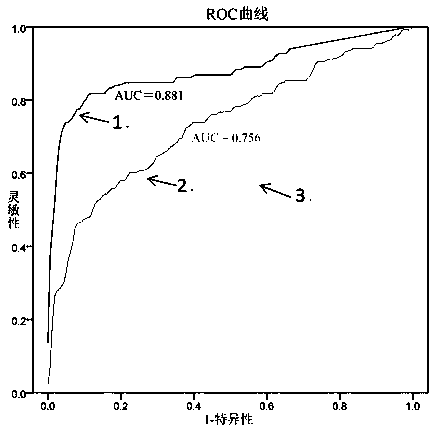
[0060] 3. According to the diagnostic results of the subject, the copy number of exosome-derived malat1, and the concentration of plasma MD-miniRNA, ROC curves of the exosome-derived malat1 and MD-miniRNA were drawn respectively by SPSS13.0 software ( see image 3 ).
[0061] 4. Conclusion: Based on image 3 The results shown show that the AUC area (0.881), sensitivity and specificity of the ROC curve of serum exosome-derived malat1 are higher than the existing marker MD-miniRNA (AUC area is 0.756),...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com