Fluorescence quantitative internal reference gene suitable for Haizhou Changshan flower, primer and application thereof
A technology of fluorescence quantification and internal reference gene, which is applied in the field of plant molecular biology, can solve the problems affecting the accuracy and instability of the target gene, and achieve the effects of strong primer specificity, improved reliability, and improved detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] 1. Plant tissue total RNA extraction and cDNA synthesis
[0029] Extract total RNA with EASYspin Plus Plant RNA Extraction Kit (Adley Biotechnology Co., Ltd.), use a spectrophotometer (METTLER TOLEDO, Switzerland) to detect RNA concentration and quality, and utilize 1.5% agarose gel electrophoresis to detect the total complete RNA. sex. Using Beijing Quanshijin Reverse Transcription Kit (Quanshijin Biotechnology Co., Ltd.), RNA was purified and the first strand of cDNA was synthesized according to the instructions.
[0030] 2. Selection of internal reference genes and design of primers
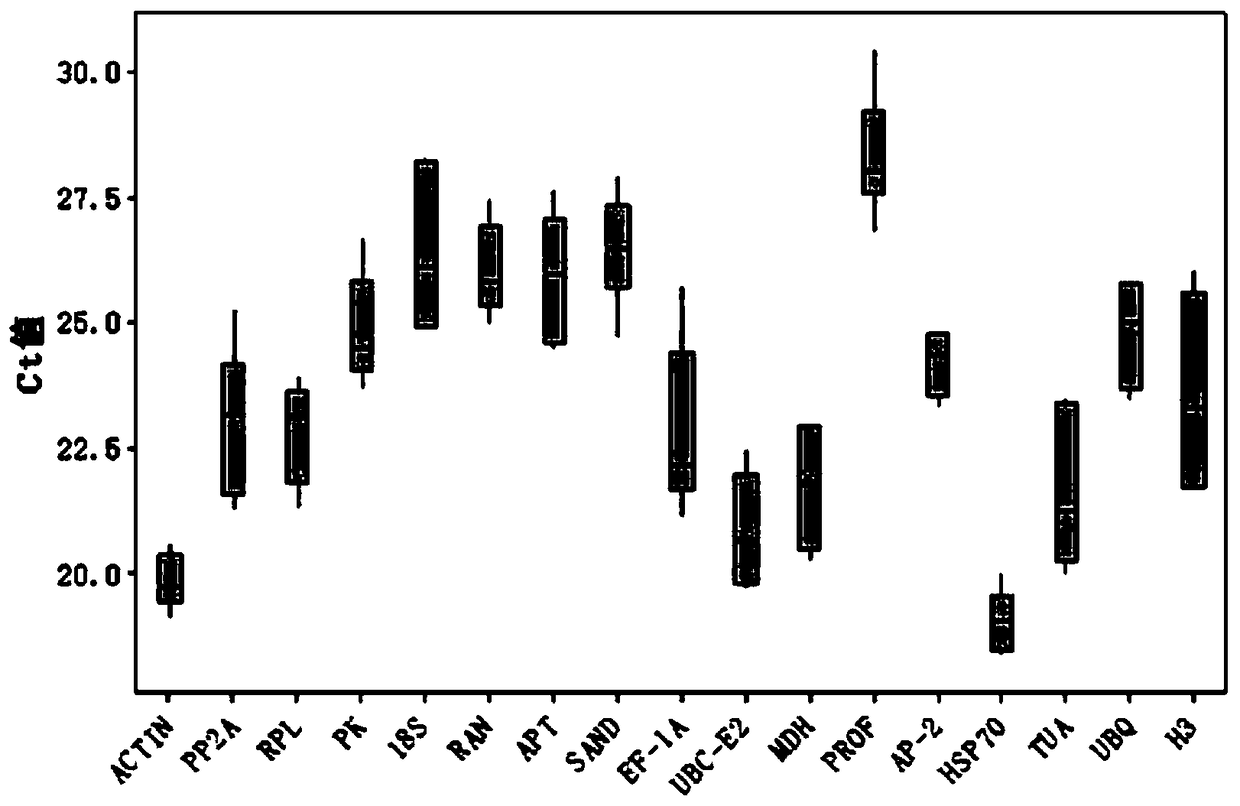
[0031] Query the main internal reference genes in other plants according to the literature, and finally design 17 internal reference genes required for the experiment, which are Actin (SEQ ID NO.1), PP2A (SEQ ID NO.2), RPL (SEQ ID NO.3 ), PK (SEQ ID NO.4), 18S (SEQ ID NO.5), RAN (SEQ ID NO.6), APT (SEQ ID NO.7), SAND (SEQ ID NO.8), PROF (SEQ ID NO. NO.9), MDH (SEQ ID NO.10), EF-1A (S...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com