Reagents, enrichment methods and applications for enrichment of highly methylated regions of dna
A high-methylation and regional technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as loss of site information, inability to meet usage requirements, and biased sequencing data
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
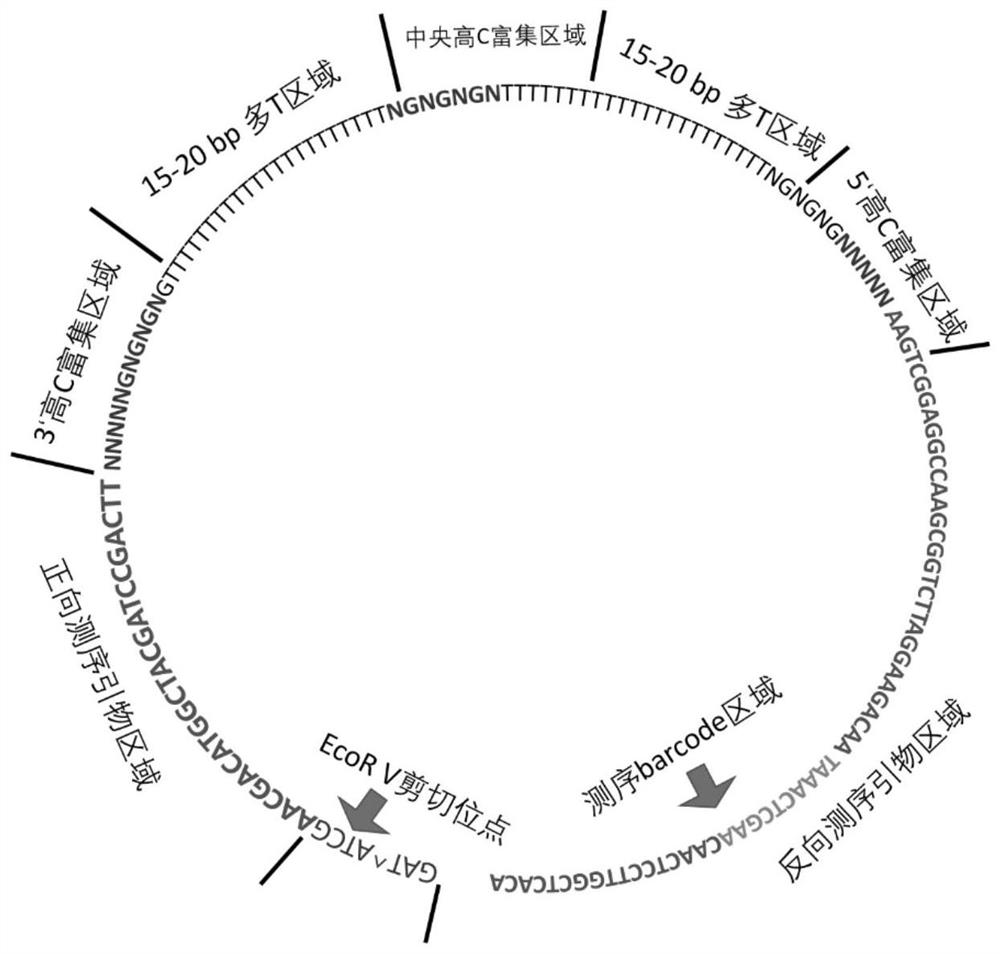
[0070] In this example, a probe was first designed based on the sequence characteristics of the hypermethylated region of free DNA, such as figure 1 As shown, the probe is a single-stranded circular structure, and the probe sequence includes an endonuclease cleavage region, a forward primer binding region, a 3' end high C fragment binding region of free DNA, a middle high C fragment binding region of free DNA, The 5' high C fragment binding region of free DNA and the reverse primer binding region, the forward primer binding region, the 3' high C fragment binding region of free DNA, the middle high C fragment binding region of free DNA, the 5 The high C fragment binding region at the 'end and the reverse primer binding region are selectively provided with a spacer between the two regions.
[0071] In the probe of this example, the primer binding region includes both the forward primer binding region and the reverse primer binding region, and the endonuclease cleavage region is ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com