DNA (deoxyribonucleic acid) probe and method for detecting DNA single base mutation
A technology of DNA probe and single-base mutation, which is applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of high professional requirements for operators, complex detection method design, and cumbersome professional operation, etc., to achieve Low professional requirements, easy operation, good effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0035] The technical solution of the present invention will be further described in detail below in conjunction with the accompanying drawings and embodiments.
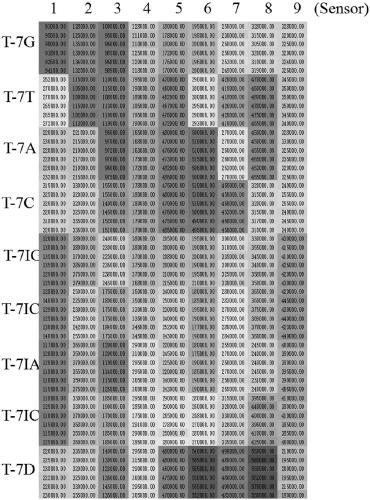
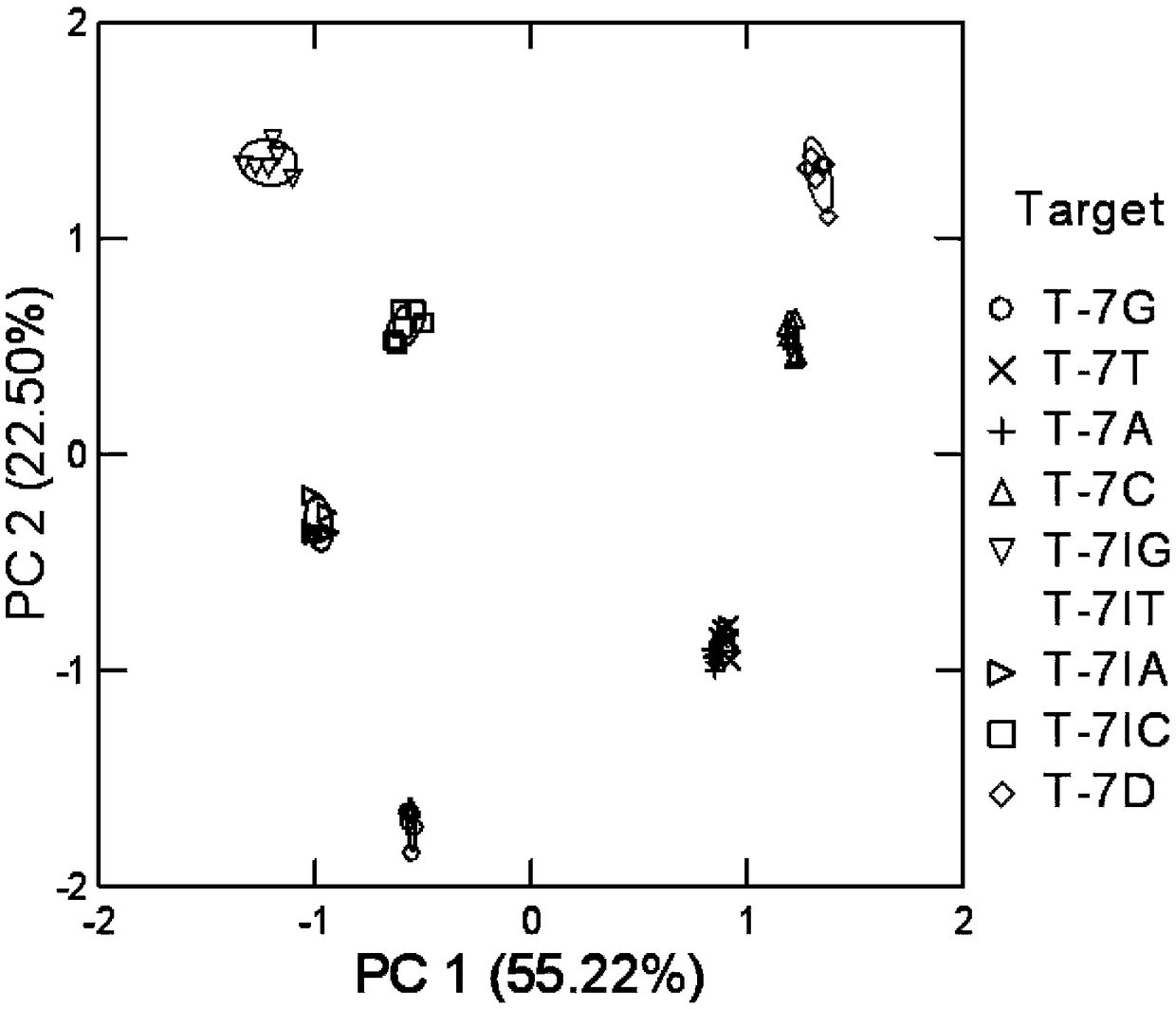
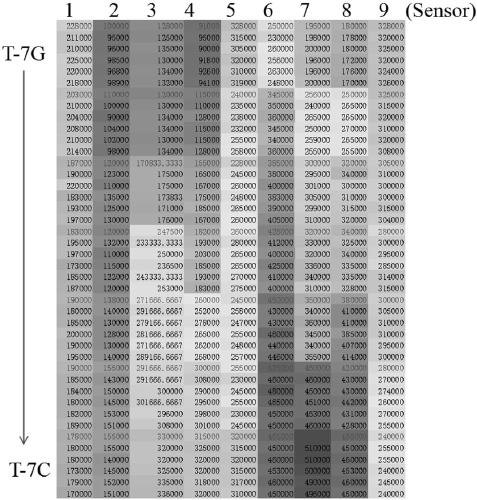
[0036] The naming of the DNA sequence of the mutation target in the embodiment represents its different base types of mismatch sites, and does not aim at specific functions, emphasizing the versatility of the method; the mixed solution of DNA of different mutation targets is obtained by artificial mixing, The discriminative ability of the method to identify different proportions of mismatches. Complex samples are obtained by adding high concentrations of protist DNA to the buffer solution to simulate the complex sample environment in actual analysis and testing, emphasizing the anti-interference ability of the method. The chemically synthesized mutation target used in the present invention is provided by Shanghai Sangong Co., Ltd. The schematic diagram of the reaction process of the detection method of the present in...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com