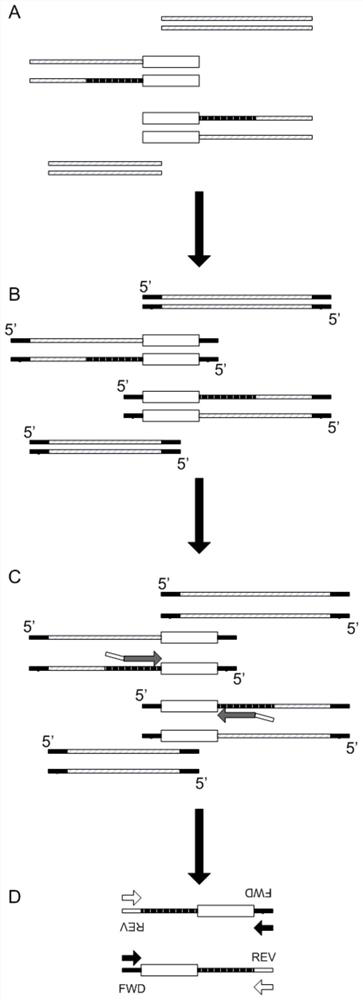
A method for detecting microbial target sequences based on single-primer probe capture
A technology of target sequence and probe sequence, which is applied in the field of microbial detection, can solve problems such as unsuitability for quantitative analysis, uneven enrichment results, and influence on result judgment, so as to eliminate inaccurate detection results, accurate and reliable detection results, and reduce The effect of testing costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] 1. Design 4 probes to capture 2 specific regions of 250bp in the Staphylococcus aureus (Sa) genome, according to figure 1 Library construction was performed using the library construction protocol shown. The part of the probe sequence used ( figure 1 Medium grey arrows) are shown in Table 2.
[0039] Table 2
[0040]
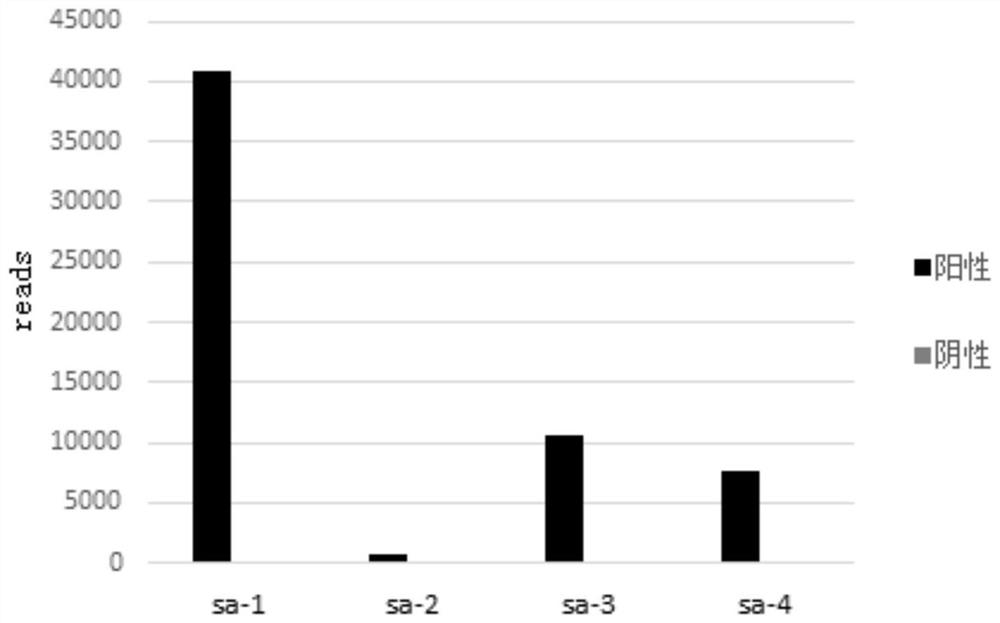
[0041] 2. The bacterial genome of Staphylococcus aureus was extracted with Tiangen DP-302 kit, and the extracted nucleic acid was quantified with Qubit dsDNA HS Assay Kit, and the genome copy number was calculated according to the genome size. Blood DNA was extracted and quantified with Tiangen Microsample DNA Extraction Kit DP-316. The equivalent of 10,000 copies of S. aureus nucleic acid was added to 10 ng of human nucleic acid.
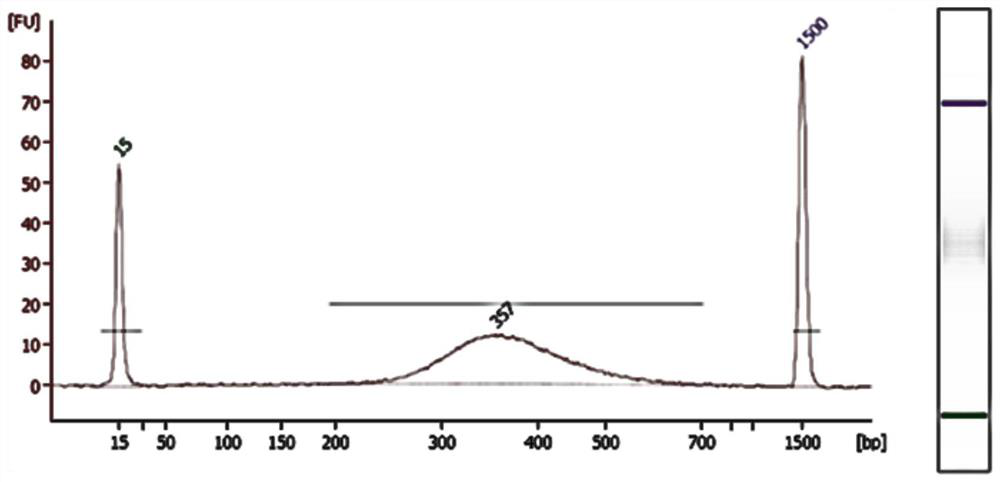
[0042] 3. Sample interruption: The sample interruption method is The Pico fragmentation method breaks the sample into fragments in the range of about 200-250bp. strictly follow To operate according to Pico's instru...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com