A composition, method and application for nucleotide sequence modification
A nucleotide sequence and composition technology, which is applied in the field of nucleotide sequence modification composition, can solve problems such as cumbersome methods, inability to target CBE, and limit the application range of single-base gene editing systems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Vector construction
[0046] (1) Construction of the first vector
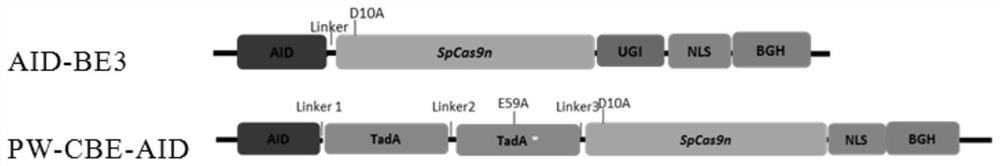
[0047] The first expression element group on the first vector is such as figure 1 shown ( figure 1 PW-CBE-AID), which includes: cytosine deaminase (AID) expression element, wild-type adenine deaminase (TadA) expression element, mutant adenine deaminase (TadAE59A, which is compared to TadA, whose amino acid residue at position 59 is mutated from E to A) expression element and mutant Cas enzyme (SpCas9n) expression element; each expression element is connected by Linker.
[0048] Wherein, the amino acid sequence of AID is shown in positions 1-182 of SEQ ID NO.6, the amino acid sequence of Linker1 is shown in positions 183-198 of SEQ ID NO.6, and the amino acid sequence of TadA is shown in SEQ ID NO.6 The amino acid sequence of Linker2 is shown in positions 365-396 of SEQ ID NO.6, the amino acid sequence of TadAE59A is shown in positions 397-562 of SEQ ID NO.6, and the amino acid sequence of Linker3 is ...
Embodiment 2
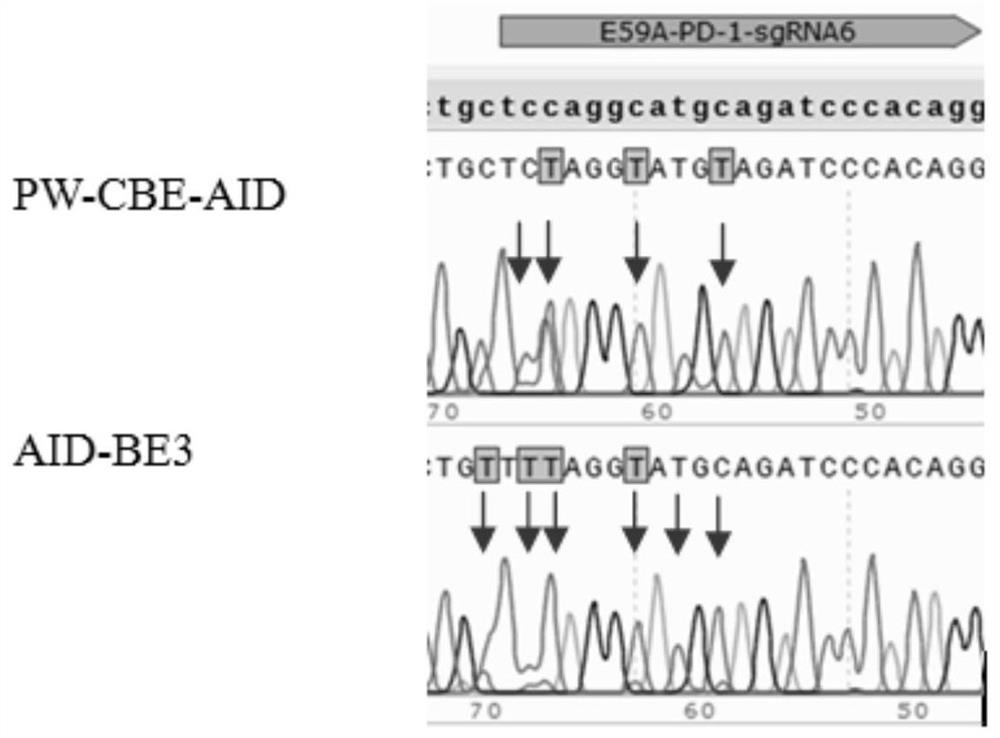
[0056] Validation of the working window for genetic modification of the composition of Example 1
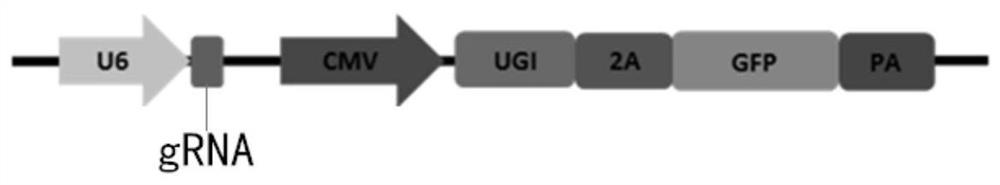
[0057] (1) Download human genes PD-1 and KCNS1 from NCBI, in which PD-1 has designed 4 targets and KCNS1 has designed 1 target (such as Table-1, the underline in the table is PAM), similar to CRISPR / Cas9 target oligo design strategy, the sgRNA uses U6 as the promoter and needs G as the transcription start site, CACC is added to the 5' end of the forward oligo for each target, and the reverse oligo is the complementary strand of the target. AAAC was added at the 5' end (see Table 2).
[0058] Table 1 Target nucleotide sequences on human gene PD-1
[0059]
[0060]
[0061] Table 2 Sequences of forward and reverse oligos for different targets
[0062] target name sequence (5`-3`) PD-1-sg6-up CACCGTCCAGGCATGCAGATCCCCAC PD-1-sg6-dn AAACGTGGGATCTGCATGCCTGGAC PD-1-sg7-up CACCGTGCAGATCCCCACAGGCGCCC PD-1-sg7-dn AAACGGGCGCCTGTGGGATCTGCAC ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com