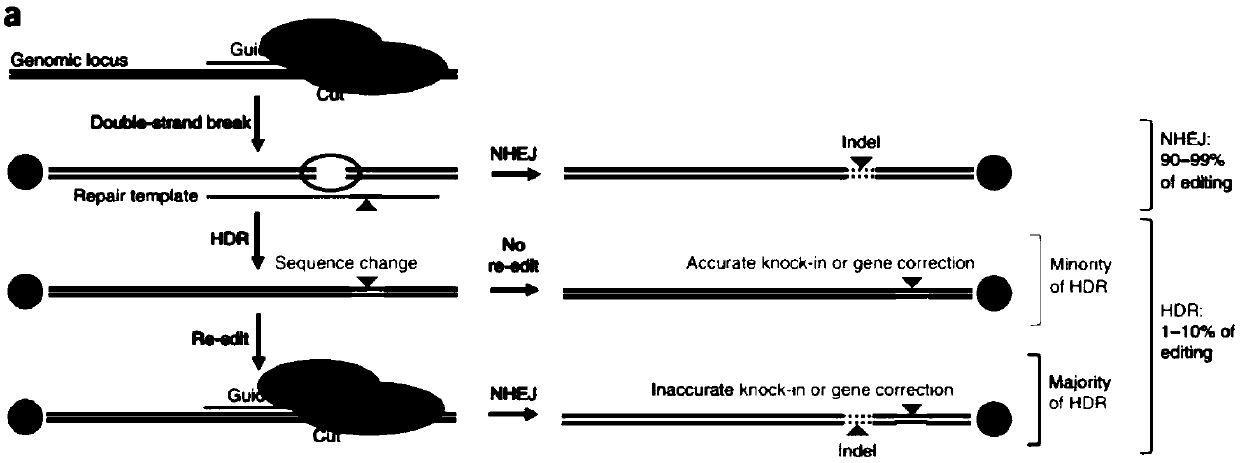
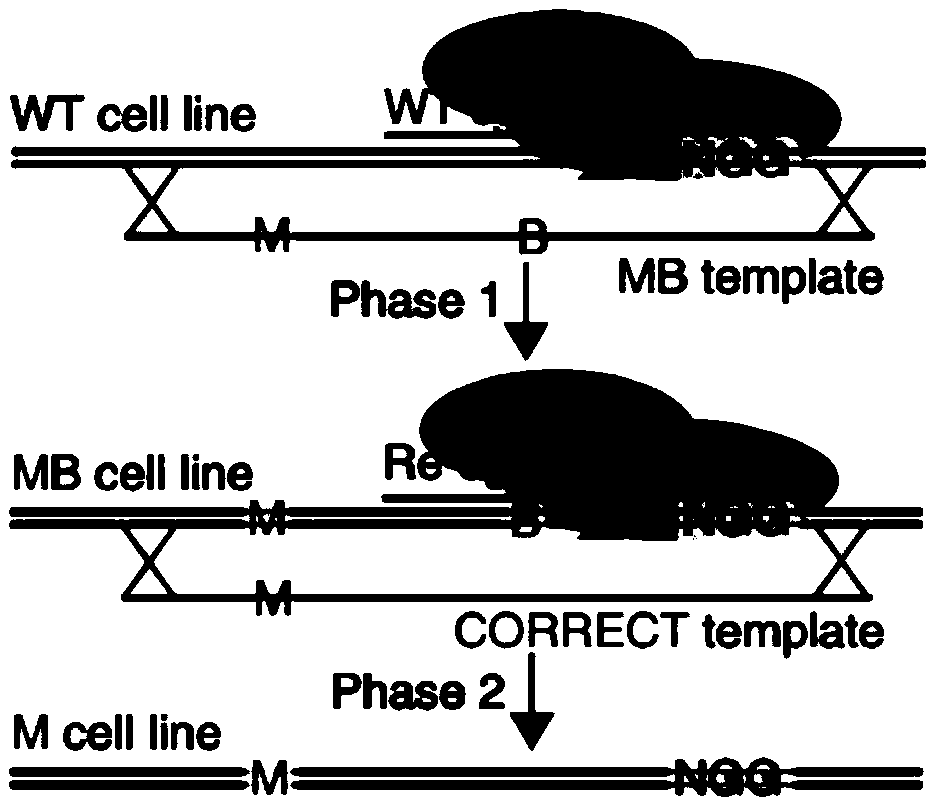
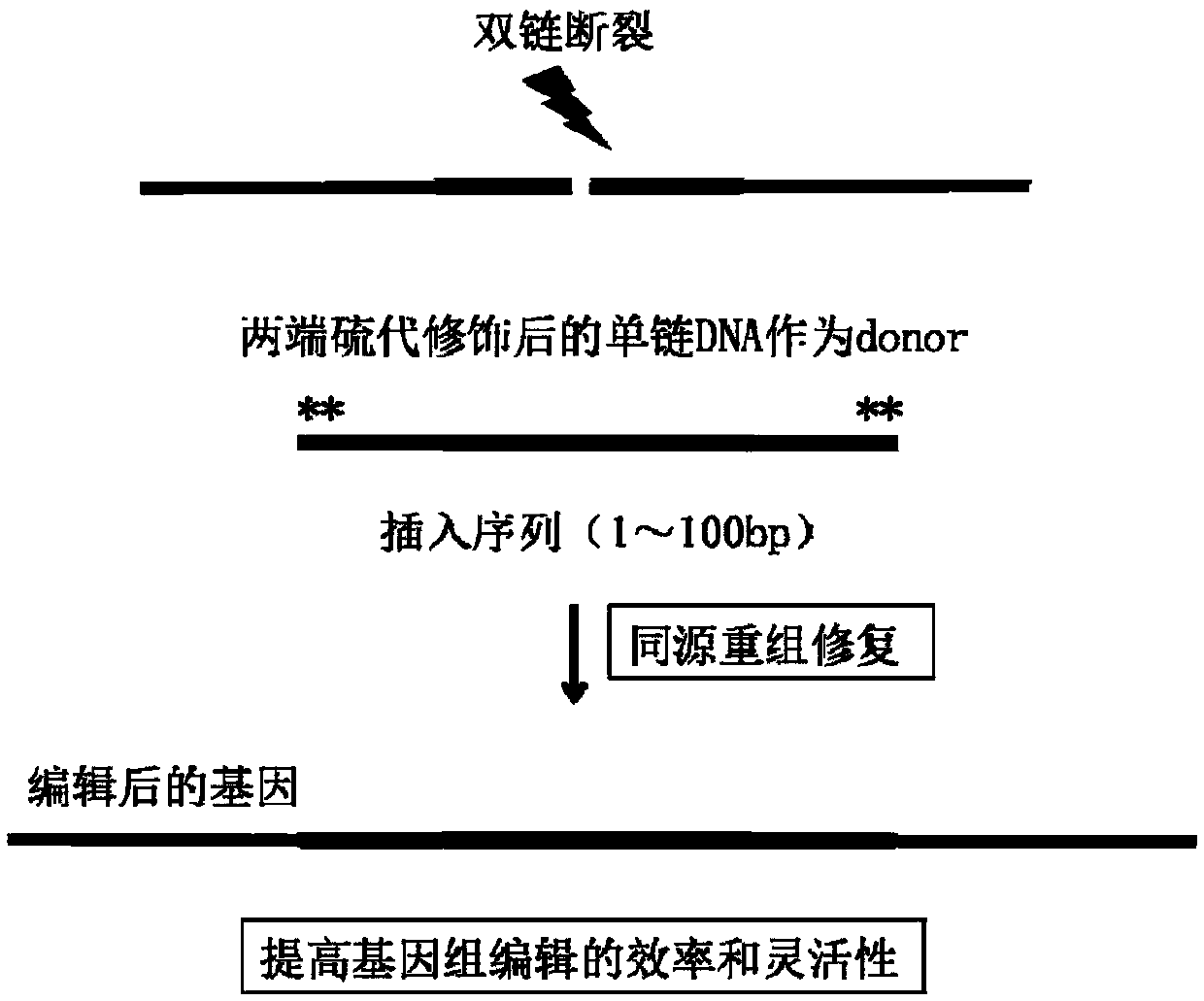
CRISPR single-base restoration system and application thereof
A repair system, single-base technology, applied in the field of gene editing, which can solve problems such as off-target, reduced stability and melting temperature, and slight toxicity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] An embodiment of the CRISPR single base repair system in the present invention, its preparation method is as follows:
[0044]The primers used in this example were designed using the Primer3 Plus online primer design tool (http: / / www.primer3plus.com / cgi-bin / dev / primer3plus.cgi); synthesized and integrated by Suzhou Jinweizhi Biotechnology Co., Ltd. The sulfur-modified donor-ssDNA was synthesized by Suzhou Jinweizhi Biotechnology Co., Ltd.; the engineering enzyme T7E1 enzyme used to identify whether the knife (that is, the CRISPR / Cas9 plasmid) cuts was purchased from NEW ENGLAND BIOLAB (article number #M0302S), and identified Donor integration repair engineering enzyme KspAI enzyme was purchased from NEW ENGLAND BIOLAB (article number is #R0105V). Sanger sequencing was entrusted to Sangon Bioengineering (Shanghai) Co., Ltd., and plasmid construction based on the sequence designed by the inventor of the present application was completed by Suzhou Jinweizhi Biotechnology C...
Embodiment 2
[0067] Example 2 Identification of the cutting effect of the CRISPR / Cas9 plasmid knife and whether the donor is integrated
[0068] identification method : After co-transfecting 293TT cells with the CRISPR / Cas9 plasmid and donor-ssDNA prepared in Example 1, extract the total cellular genomic DNA and carry out Sanger sequencing to verify the cutting effect of the CRISPR / Cas9 plasmid knife and whether the donor is integrated.
[0069] The specific operation method is as follows:
[0070] (1) Cell culture
[0071] The 293TT cell line was incubated with complete DMEM medium containing 10% serum at 37°C, 5% CO 2 Cultivated in an incubator. When the cell confluency reached 90%, it was digested with 0.25% trypsin, then terminated with DMEM complete medium, inoculated into a 6-well plate, and continued to culture for 24 hours.
[0072] (2) Plasmid transfection
[0073] After 24 hours, confirm that the cells are well adhered to the wall and the cell confluency reaches 80%, and th...
Embodiment 3
[0112] Example 3 Verification of site-directed mutagenesis effect of the CRISPR single base repair system of the present invention
[0113] identification method : By selecting monoclonal cells to further identify and screen out the 293TT cell line with β17 mutation and nonsense mutation; specifically, each group of cells after transfection 48h in Example 2 was digested with 0.25% trypsin and blown into a single Cells were suspended in DMEM medium with 10% fetal bovine serum for later use.
[0114] Dilute the cell suspension in gradient multiples, each group of cells were respectively inoculated in a dish containing 10mL 37°C pre-warmed culture medium at a gradient density of 50, 100, and 200 cells per dish, and rotated gently to make the cells evenly dispersed. Ultimately, the culture wells contain at most 1 cell. Set at 37°C 5% CO 2 and cultured in a cell culture incubator with saturated humidity for 2-3 weeks.
[0115] Observe frequently, when there are clones visible ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com