Prediction method for ubiquitination degradation of target protein
A ubiquitination and protein technology, which is applied in proteomics, instruments, biological neural network models, etc., to achieve low cost and improve the effect of transformation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
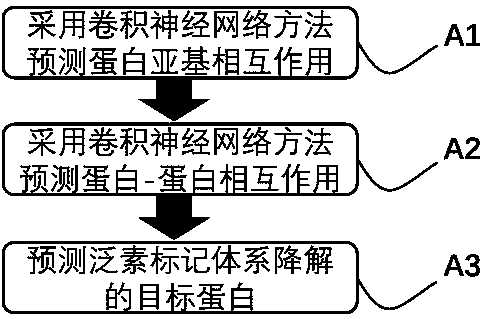
[0042] This embodiment provides a method for predicting ubiquitination and degradation of the signal transduction protein Smad3, comprising the following steps:
[0043] A1. Using convolutional neural network method to predict protein-protein interaction, including the following steps:
[0044] Step1: Select the three-dimensional structure of Homo sapiens from the PDB protein database, and the three-dimensional structure resolution of the protein-protein complex is less than or equal to 3 angstroms, and download the pdb or cif file, and then use the YASARA software python programming to intercept the protein atoms contained in the pdb or cif file The data of the three-dimensional coordinates is saved as a new pbd or cif file;
[0045] Step2: Use the python programming of YASARA software to calculate the type and frequency of protein-protein interaction sites and amino acid residues on the interface in the three-dimensional structure of the protein-protein complex obtained in t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com