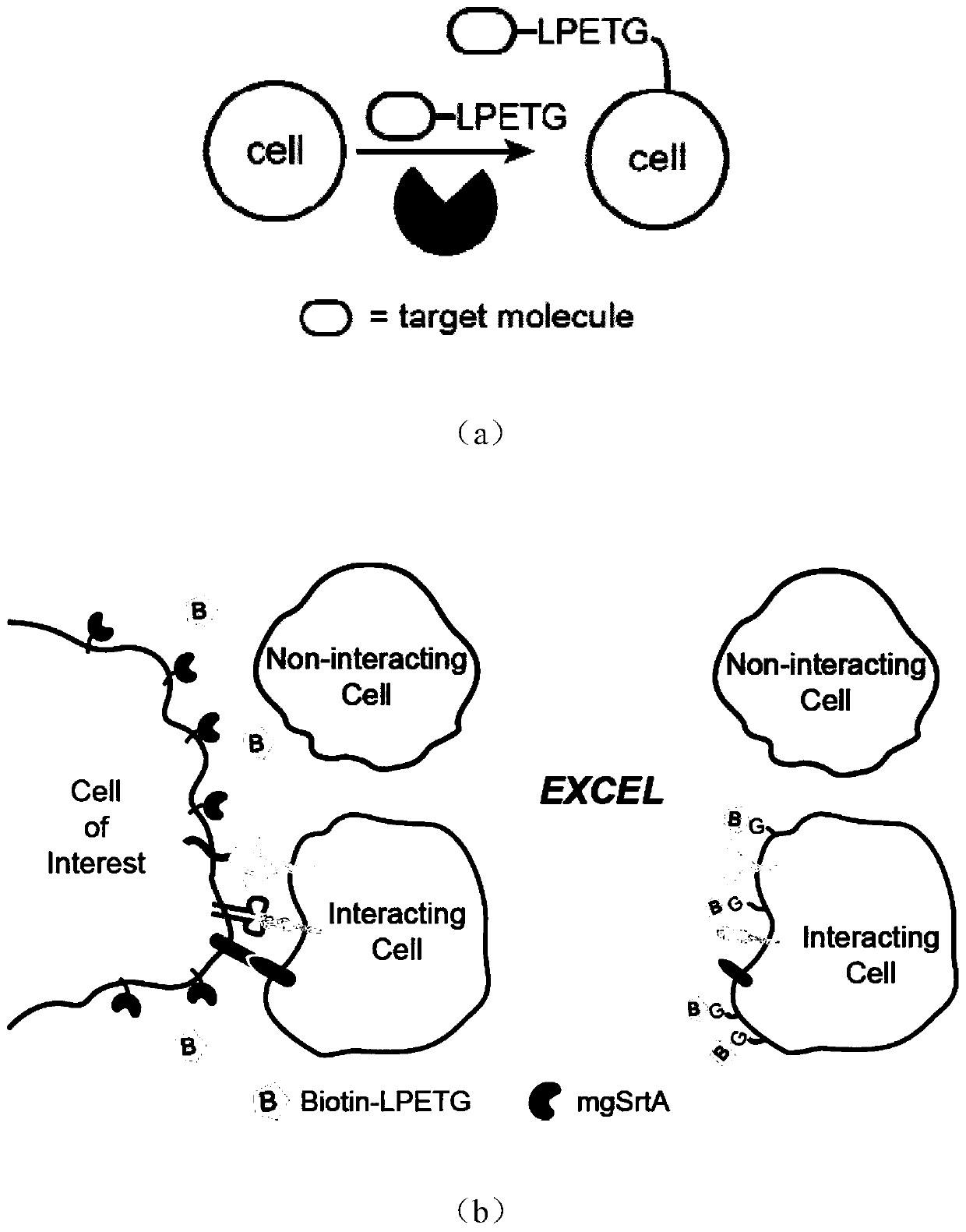
Enzyme and method for marking cell membrane surfaces and researching cell-cell interaction
A labeling method and cell surface technology, applied in the field of bioengineering, can solve the problem of low reactivity, achieve precise capture and labeling, and improve labeling efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
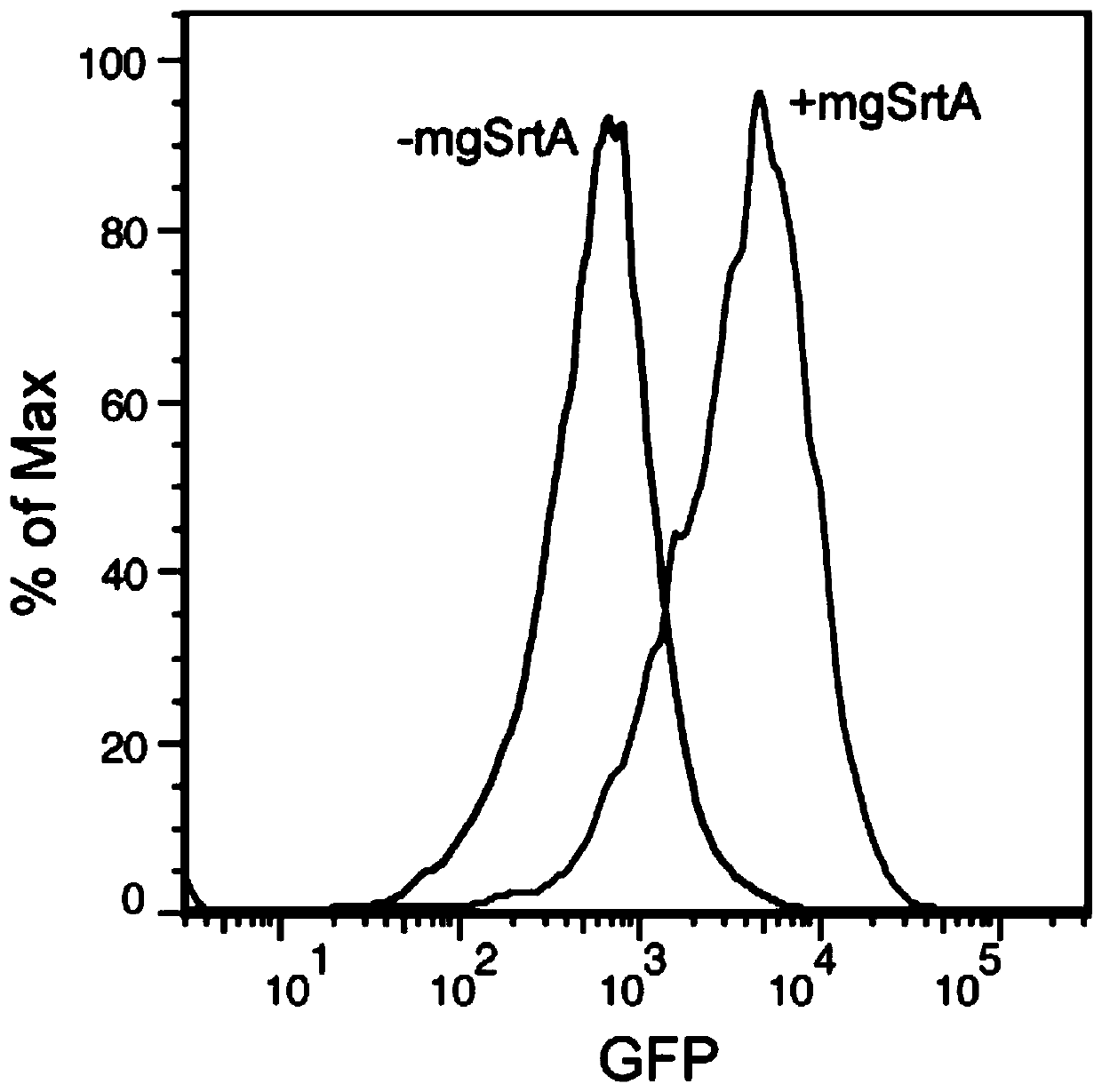
[0037] Example 1, Cell Membrane Labeling
[0038] The eGFP-LPETG and the Staphylococcus aureus sortase A mutant involved in the present invention were simultaneously expressed on the pet28a vector by a cloning method, and the protein was purified using a 6×His tag. Cell membrane labeling reaction was performed in PBS buffer or Tris buffer (30mM Tris, 150mM NaCl, 5mM CaCl 2 , pH 7.4), the reaction conditions are as follows: 20 μM mgSrtA, 20 μM eGFP-LPETG, room temperature for 1 hour, cell density 2×10 7 cells / mL. After the reaction was completed, the buffer was used to wash three times, and then the flow cytometer was used for characterization. The result is as figure 2 As shown, it can be seen that the fluorescence intensity of eGFP in cells is stronger after adding mgSrtA (+mgSrtA) compared with adding only eGFP-LPETG (-mgSrtA), which proves that eGFP-LPETG is labeled on the cell surface. The experimental results of the six mutants shown in SEQ ID No: 1 to 6 in the seque...
Embodiment 2
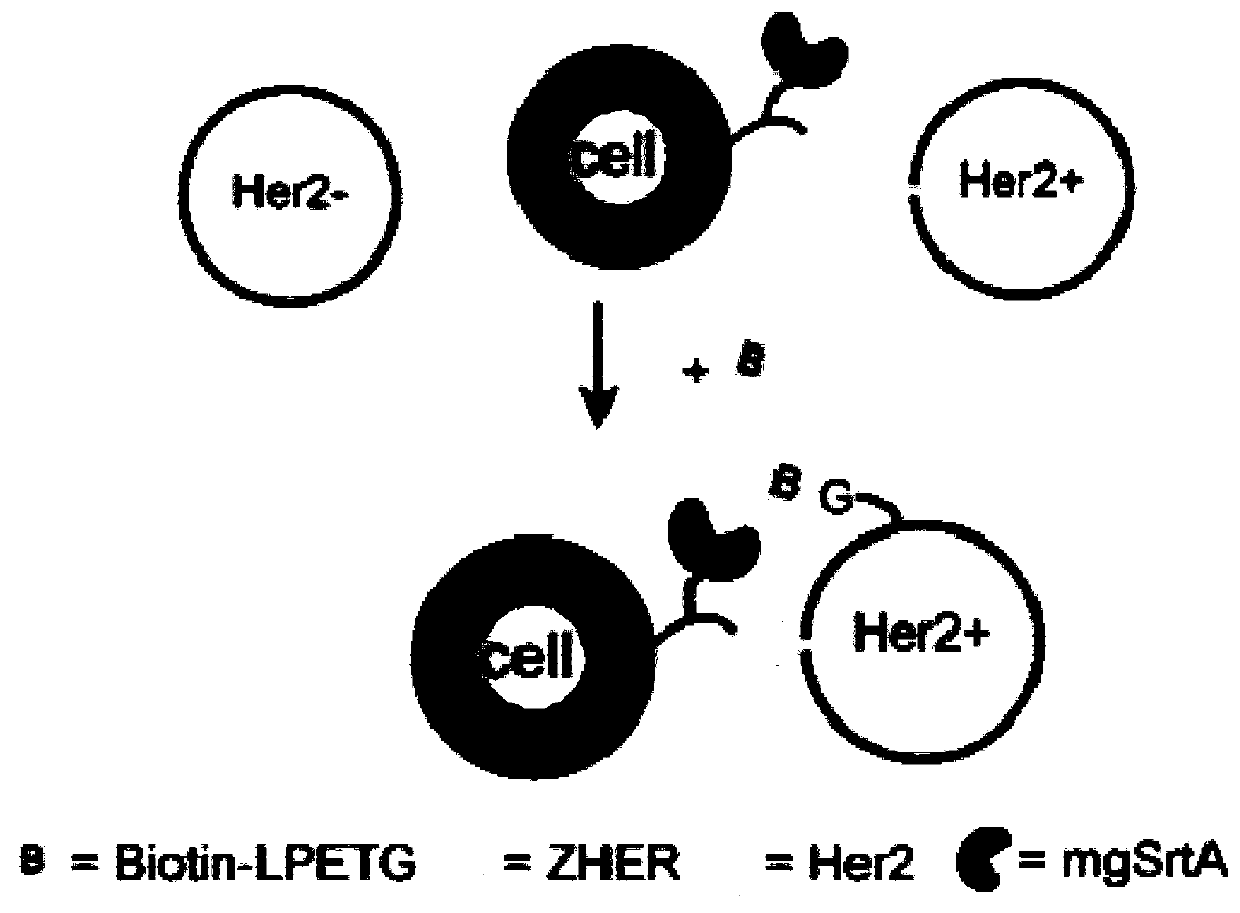
[0039] Example 2. Recording and detection of model HER2 positive cell-cell interaction
[0040] First, ZHER and / or mgSrtA were displayed on the surface of HEK293T cells by genetic modification, and eFluor670 was labeled as a signal for later sorting. MDA-MB-231 cells were stably transfected with HER2 receptor (human epidermal growth factor receptor-2) by lentiviral transfection as Her2 positive cells (Her2+ cells). The above two kinds of cells (cell density 2×10 7 cells / mL) were co-incubated in PBS buffer at room temperature for 30 minutes, and 100 μM biotin-LPETG was added for labeling, and MDA-MB-231 cells themselves were Her2-negative cells (Her2-cells) as a control. After labeling, wash with PBS three times, then incubate with Streptavidin PE on ice for 15 min, and finally use flow cytometry to characterize.
[0041] In this experiment, Her2-positive cells interacted with cells expressing the Her2 ligand ZHER, allowing mgSrtA to label biotin on the surface of Her2-positi...
Embodiment 3
[0042] Example 3. Detection of model Raji cell-cell interaction
[0043] First, CD40L (or CD40L* or CTLA4) and mgSrtA are displayed on the surface of HEK293T cells by genetic modification, and cells expressing CD40L or CTLA4 can induce the interaction between HEK293T and Raji cells by pairing with CD40 or B7 on the surface of Raji cells , CD40L* is a mutant of CD40L that does not interact with CD40L and serves as a negative control. The above-mentioned HEK293T cells expressing different ligands were co-incubated with Raji cells (Raji cells were pre-labeled with eFluor670) in PBS buffer at room temperature for 30 minutes, and at the same time, 100 μM biotin-LPETG was added for labeling. After labeling, wash with PBS three times, then incubate with Streptavidin PE on ice for 15 min, and finally use flow cytometry to characterize.
[0044] In this experiment, the interaction between cells expressing CD40L, CD40L* or CTLA4 and Raji cells expressing CD40 and B7 allowed mgSrtA to l...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com