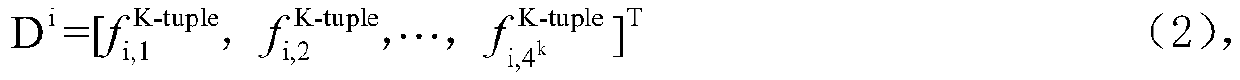Method for predicting interaction between sRNA and target mRNA of sRNA in bacteria
A prediction method and target technology, applied in instrumentation, genomics, proteomics, etc., can solve problems such as dependence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
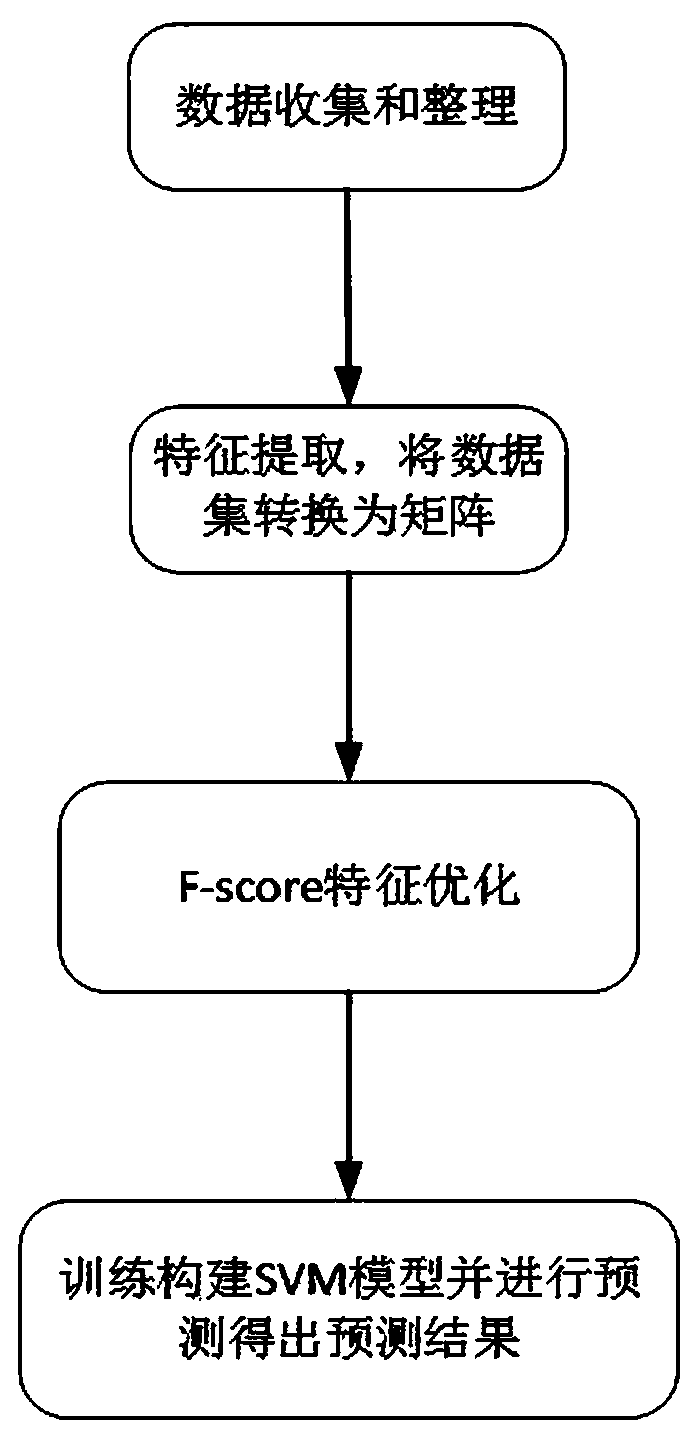
[0049] refer to figure 1 , a method for predicting the interaction between sRNA and its target mRNA in bacteria, comprising the steps of:
[0050] 1) Data collection and arrangement: Obtain the sRNA-mRNA interaction data set from the sRNATarBase 3.0 database, compare the mRNA primary sequence in the data set with the corresponding mRNA whole genome sequence in the NCBI database, and intercept the mRNA upstream of the start codon The sequence fragment between 80nt and downstream 50nt, the start codon is AUG, and then, connect the sRNA sequence and the mRNA sequence to form a sequence pair, each sequence pair is composed of one sRNA sequence and one mRNA sequence, such as: sRNA -LLLLLL-mRNA, where L is the connection symbol, and the curated data set includes 241 positive sample RNA sequence pairs with interactions and 185 negative sample RNA sequence pairs without interaction;
[0051] 2) feature extraction, data set is converted into matrix: its process is:
[0052] Suppose t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com