Rice genome recombinant nucleic acid fragment RecCR012612 and detection method thereof
A technology for recombinant nucleic acid and genome, applied in the field of recombinant nucleic acid fragments and its detection, can solve the problems of long time consumption and low efficiency, and achieve the effect of improving rice blast resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1 Breeding of recombinant plants with gene fragments resistant to rice blast
[0043] The materials used in this embodiment are rice'Y58S' and rice'Gumei 4'.
[0044] Rice'Gumei 4'has good resistance to rice blast, and it is speculated that the Pi2 interval of chromosome 6 played a key role in the resistance of this material to rice blast.
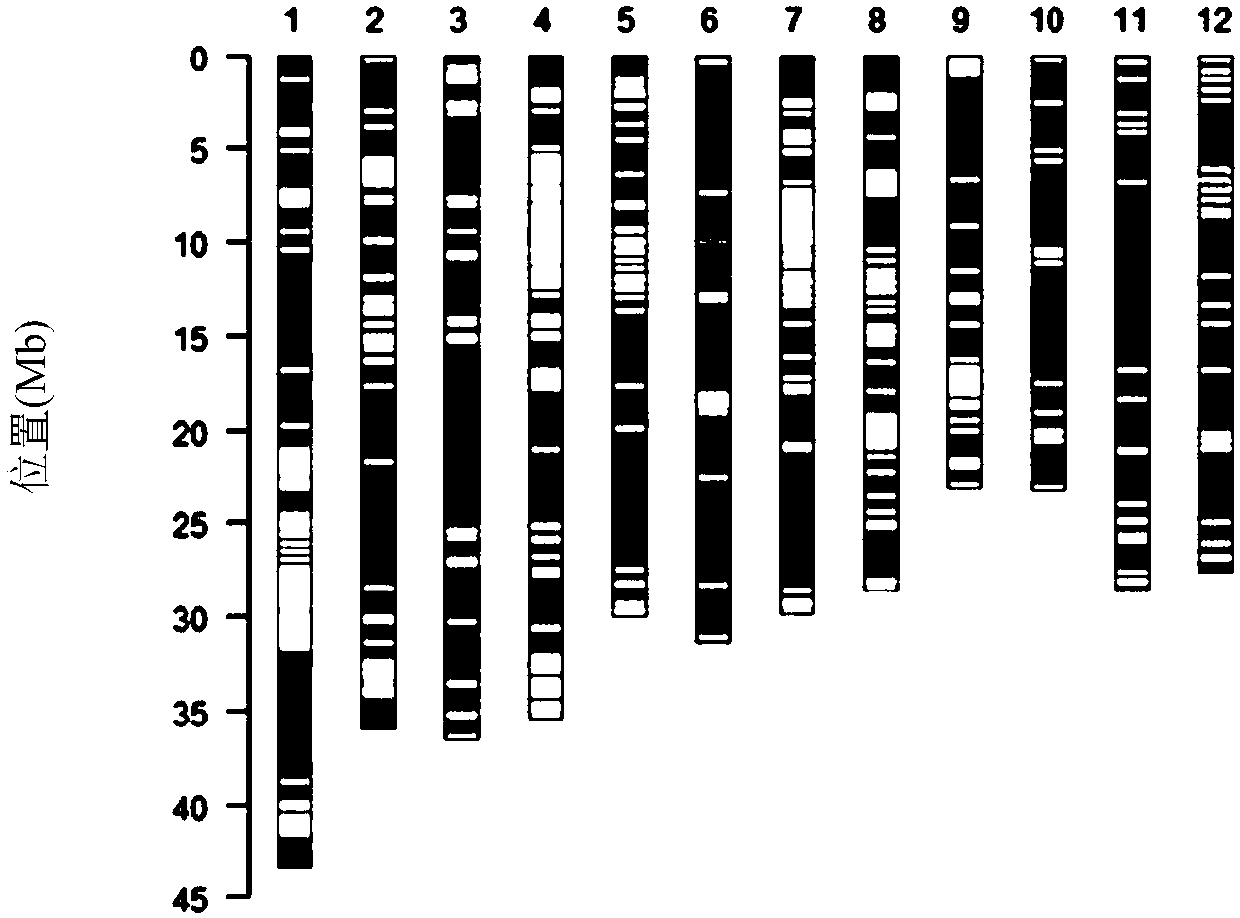
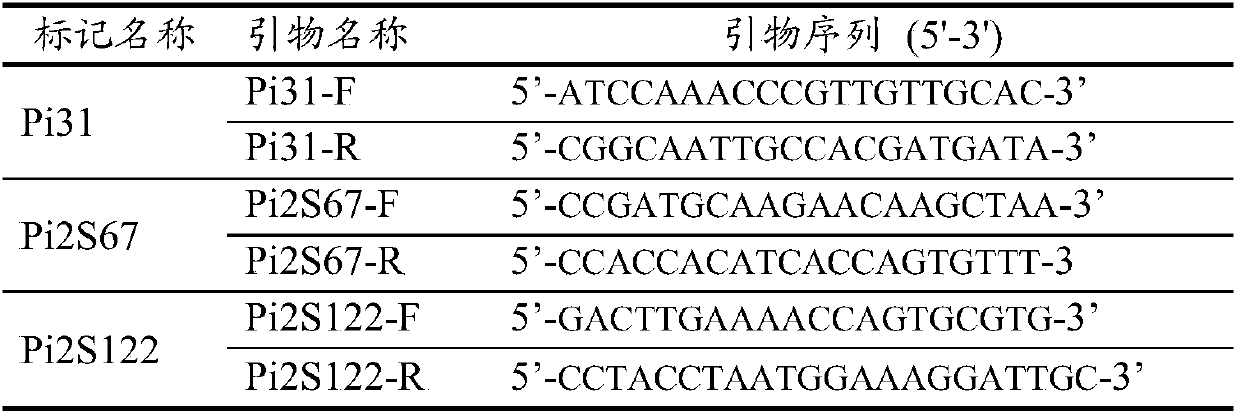
[0045] In the process of selection of recombinant plants, molecular markers were used to perform prospect selection on recombinant plants, and the used prospect selection molecular markers were screened. Refer to the 6.1 edition of the MSU / TIGR annotation of the rice Nipponbare genome to download the 9,559,000 to 10,990,000 DNA sequence of chromosome 6. Use SSRLocator software to scan the SSR sites in the above sequence. Primer Premier 3.0 software was used to design primers for the found SSR sites, and a total of 162 pairs of primers were designed. The PCR method was used to screen the polymorphisms of the above primer pairs i...
Embodiment 2
[0055] Example 2 Determination of homologous recombination fragments after introduction of rice blast resistance genome fragments
[0056] In order to determine the size of the introduced rice blast resistance genome fragment, the homozygous single plant of the'Y58S' introduced fragment was sequenced for the homologous recombination fragments on both sides of the target genome fragment. The recombinant nucleic acid fragment of the rice blast resistant genome contained in CR012612 was named RecCR012612.
[0057] Preliminarily confirmed by the detection results of the rice whole genome breeding chip RICE60K, the upstream homologous recombination fragment of RecCR012612 is located between the markers R0610187834CT and F0610202034AC, and the downstream homologous recombination fragment is located between the markers F0610692034AG and R0610699627AG.
[0058] At the same time, the Miseq sequencing technology was used to perform whole-genome sequencing on three samples of'Y58S','Gumei No. ...
Embodiment 3
[0071] Example 3 Resistance identification of ‘Y58S’ after introducing the rice blast resistance genome fragment
[0072] In order to identify the resistance effect, the new line CR012612, the recurrent parent'Y58S', the rice blast resistant variety Gumei 4 (as a positive control), and the rice blast susceptible variety Lijiang Xintuan Heigu (as a negative Control) was planted indoors, and then cultivated to the 3-4 leaf stage for identification using the following methods:
[0073] Select M15Bb-1-1, M15Bb-1-2, M15Bb-2-1, M15Bb-3-1, M15Bb-4-1, M15Bb-5-1, M15Bb-6-1 separated from Yichang in 2015. 7 Two strains were used as inoculation strains. Strains are stored at -20°C using the sorghum grain method. Before use, remove the stored sorghum grains to a potato dextrose medium (PDA) plate for activation (PDA: peeled potatoes 200g, glucose 20g, agar powder 15g, distilled water to make the volume to 1L), After 5 days of light cultivation at 28°C, take a fresh mycelial block with a di...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com