Method for isolating highly pure nucleic acid with magnetic particles
A magnetic particle and nucleic acid technology, applied in the preparation of test samples, DNA preparation, recombinant DNA technology, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1 and 2
[0105] Examples 1 and 2: Purification of DNA:
[0106] DNA from bacterial cells was isolated according to the method of the invention (Example 2) and the method of the prior art (Example 1). Conditions and working solutions are shown in Table 1 below. use the trademark for (Qiagen, Hilden, Germany; Cat. No. 51304) Commercial kits were used for lysis of E. coli or whole blood cells according to standard protocols. use The kit uses a magnetic bead extraction platform (trademark Gilson Gilson Inc., Middleton, US / SalusDiscovery) was used to purify DNA using magnetic particles. In the standard method of Example 1, DNA is bound to magnetic silica particles from a buffer containing a high concentration of chaotropic salts to establish binding conditions. Applying a magnetic field separates the magnetic particles from the sample solution. In particular, the magnetic particles are drawn out of solution and the sample solution is subsequently removed. In lysis step 1 (table)...
Embodiment 3
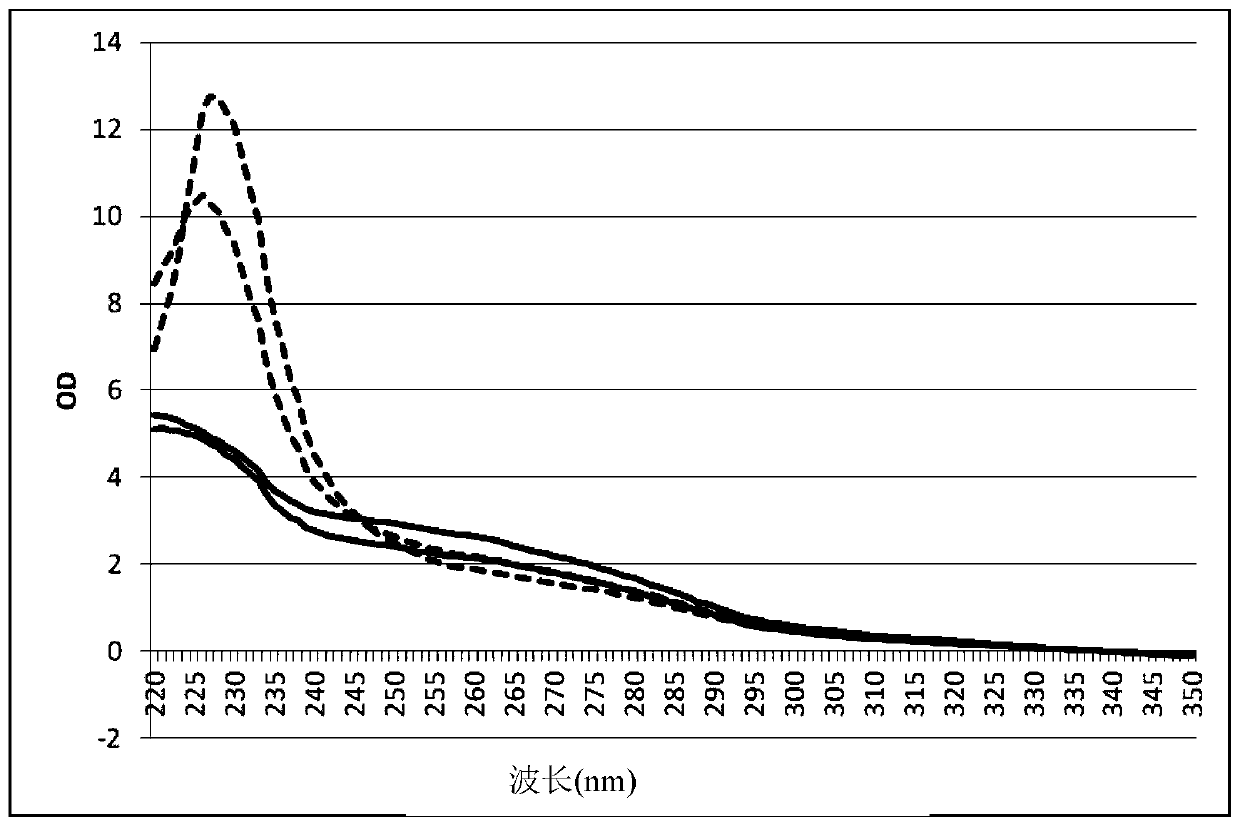
[0114] Example 3: Purity of genomic DNA from E. coli
[0115] The impurities in the Escherichia coli genomic DNA prepared in Examples 1 and 2 above were determined by spectrophotometry. To wash away contaminants from the magnetic particles, the magnetic particles were eluted by placing them in a magnetic stirrer for three minutes. The eluate was analyzed for impurities with a spectrophotometer at 220nm-350nm, where most salt residues were determined. The results are shown in figure 1 . The dotted line shows the values obtained for the detection of DNA isolated according to the water-rinsing protocol of Example 1 with the two probes (comparison). The solid line shows the values obtained for DNA isolated according to the water-rinse protocol of Example 2 detected with both probes. High OD values between 220-245nm usually indicate salt and other contaminants. The results show that the inventive method of washing the DNA-bound magnetic particles with distilled water ...
Embodiment 4
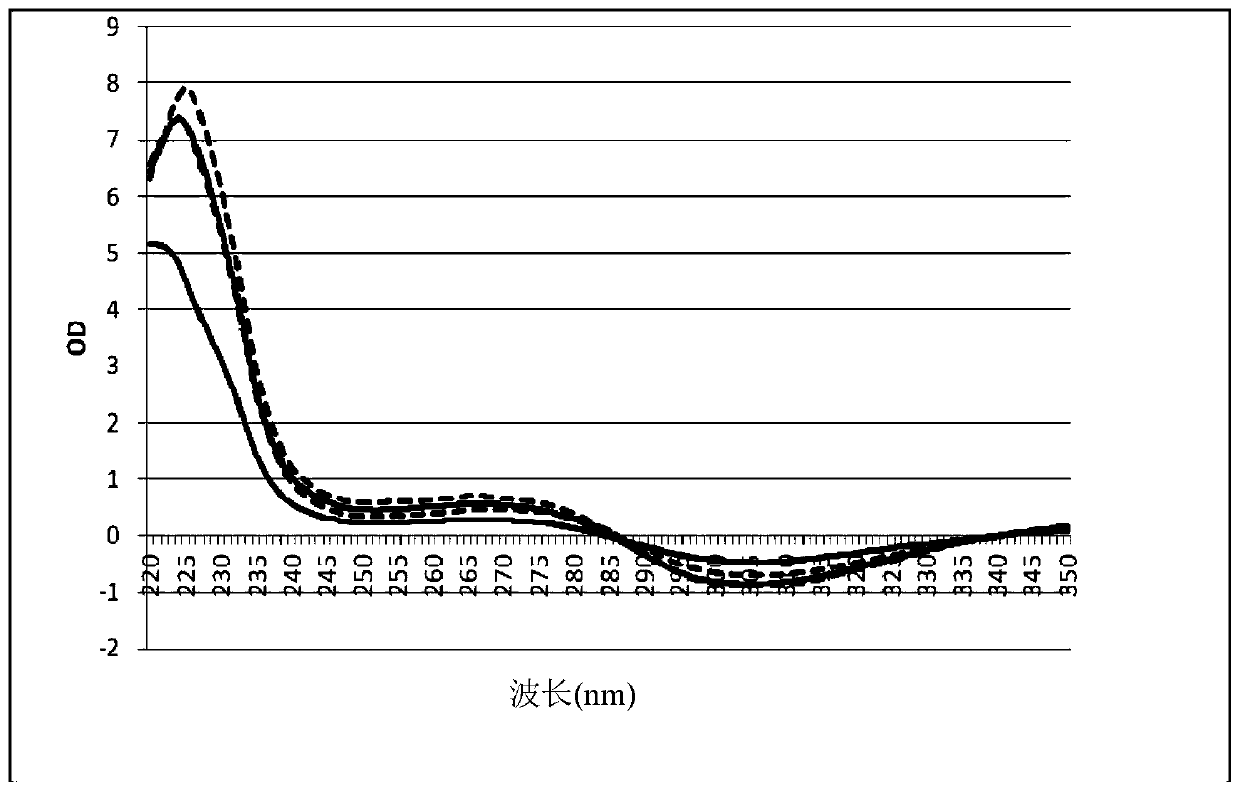
[0116] Example 4: Purity of DNA from blood
[0117] DNA from human whole blood was prepared according to the protocols of Inventive Example 2 and a modified Comparative Example 1. In the method of Example 1 above, the third wash step of the water-rinse step 5a (see table) was replaced with PE buffer. During the first and second PE buffer wash steps 3, 4 and the third PE buffer wash step, the magnetic particles fell into solution three times. The purity of the isolated DNA products was compared. The goal was to determine whether the additional washing step in the comparative method (3 drops into ethanol buffer PE) could remove impurities more efficiently than the inventive method in which the particles were only dropped into water once.
[0118] The results are shown in figure 2 . Dashed lines indicate DNA isolated by a comparative triple wash method. The solid line indicates the DNA of Example 2 isolated by the water-drop method. The results show that the method of th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com