Hierarchical TADs differential analysis method in Hi-C contact matrix based on online machine learning
A technology of machine learning and difference analysis, applied in the biological field, can solve problems affecting the recognition rate of differential TADs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0039] The implementation of the present invention will be described in detail below in conjunction with the drawings and examples.
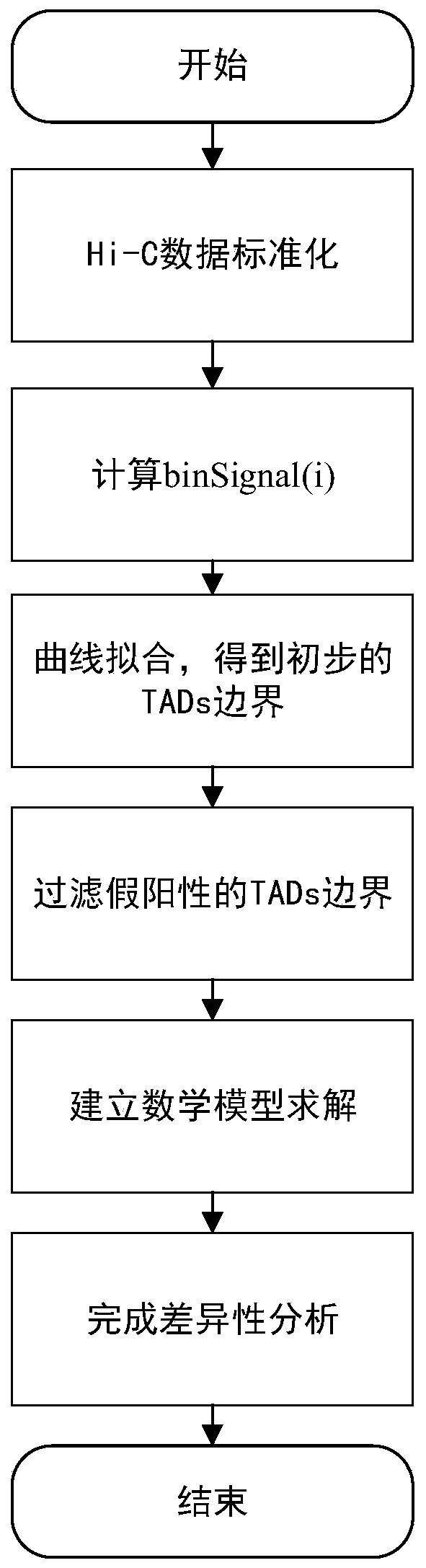
[0040] Such as figure 1 As shown, the difference analysis of hierarchical TADs under the two cell lines based on TADs boundary region recognition of the present invention comprises the following steps:
[0041] Step 1. Standardize the Hi-C data to eliminate the systematic deviation of the Hi-C experiment and enhance the comparability between the data. The specific method is as follows:
[0042] Firstly, multiHiCcompare, a cross-cell line Hi-C data normalization method, was used to preliminarily process the Hi-C data under different cell lines, so as to eliminate the systematic deviation between different cell lines as much as possible; then, the data standardization method CPM (Counts per million) to process the preliminary processed Hi-C data to further enhance the comparability of Hi-C data among different cell lines.
[0043] Step 2. Calcul...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com