Tetraploid elytrigia elongata 3E chromosome molecular marker and application thereof
A technology of molecular marker, ryegrass, applied in the field of crop genetics and breeding
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Example 1 Based on GBS technology to obtain tetraploid E. elongatum 3E chromosome-specific fragments
[0059] When the materials such as diploid E. elongatum, tetraploid E. elongatum, parent 8801 and common wheat-tetraploid E. elongatum 1-7E chromosome substitution line grow to the seedling stage, take the leaves and send them to Beijing Novogene Technology Co., Ltd. carried out GBS sequencing, and obtained the effective sequence of each sample through GBS sequencing.
[0060] The sequence of the obtained wheat-tetraploid E. elongatum 3E / 3D substitution line was first compared with the known sequence of Chinese spring, and the sequence with a similarity of 22.85% (23%) or more to wheat was eliminated, and the remaining The following sequence was then compared with the sequence of 8801 and tetraploid E. elongatum to obtain a sequence with a similarity greater than 22.85% (23%). The remaining sequences were then compared with diploid E. elongatum and other substitution li...
Embodiment 2 4
[0061] Example 2 Development of Chromosome-Specific Molecular Markers of Tetraploid E. elongatum 3E
[0062] Primers were designed for the 269 specific sequences obtained above. The PCR primers were designed using the online software Primer3 Plus, and all the primers were synthesized at Chengdu Qingke Weiye Biotechnology Co., Ltd.
[0063]The PCR reaction system is as follows: the total volume of the PCR reaction is 25uL, DNA template (concentration needs to be diluted to about 100ng / uL) plus 1uL, upper and lower primers 1uL (10uM), 2×TaqMasterMix (P114) 12.5uL, ddH 2 O9.5uL.
[0064] The PCR program was: 94°C for 5h; 94°C for 30min, 50-60°C for 30min, 72°C for 2h, 35 cycles; 72°C for 10h; 12°C.
Embodiment 3
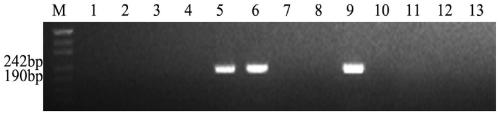
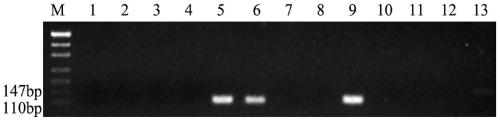
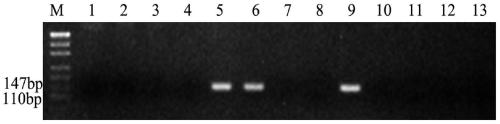
[0065] Embodiment 3 agarose gel electrophoresis detection
[0066] The PCR products were detected by 3% agarose gel electrophoresis, and the designed primers could amplify specific bands in tetraploid Echinopsis elongatum, parent 8801, and 3E / 3D substitution lines, and in CS, Shumai 482, Chuannong 16, diploid E. elongatum, 1E / 1D substitution line, 2E / 2A substitution line, 4E / 4D substitution line, 5E / 5D substitution line, 6E / 6D substitution line, 7E / 7D substitution line No bands could be amplified in the substitution lines, and this primer was the specific primer for the 3E / 3D substitution line, that is, the specific molecular marker for the 3E chromosome of tetraploid E. elongatum. 246 pairs of primers were successfully designed, and 146 pairs of specific primers were obtained after PCR amplification, product recovery, and sequencing verification in the above materials, and the molecular marker development efficiency reached 59.35%. The primer sequences of 20 pairs of specifi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com