Method and kit for enriching circulating tumor DNA on basis of multiplex PCR
A kit and tumor technology, applied in the direction of tumor/cancer cells, biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of sample loss, sensitivity reduction, etc., to reduce errors, reduce false positives, and reduce costs. loss effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
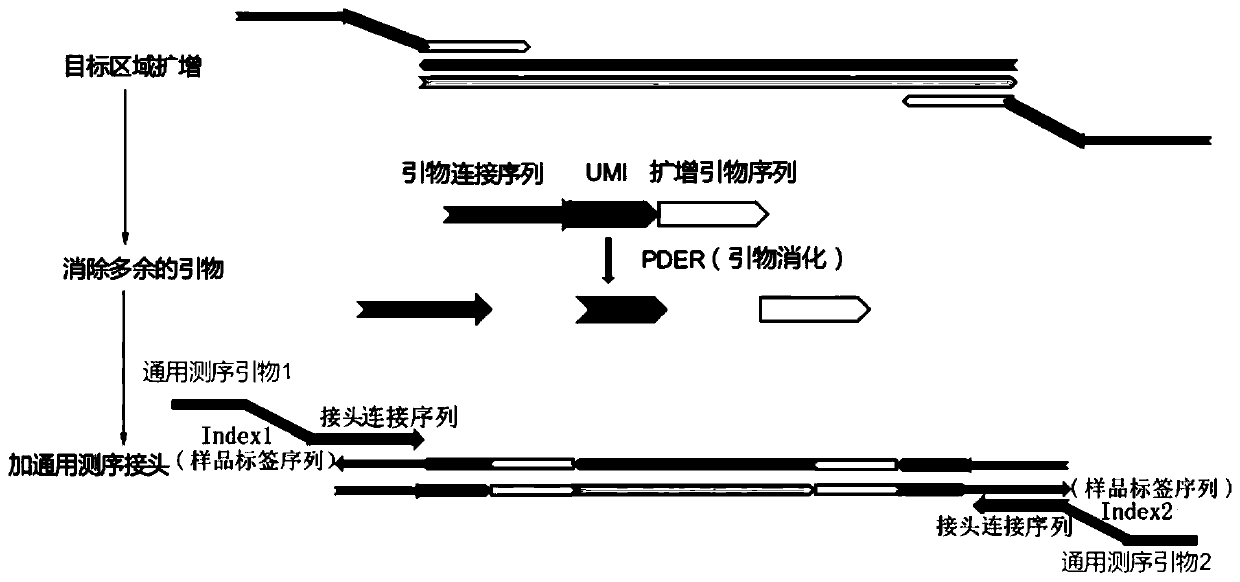
[0096] The method of the invention is used to detect ctDNA tumor mutations in blood samples.
[0097] Specific steps are as follows:
[0098] 1. Test template preparation.
[0099] The cultured SK cell line and HCC1975 (containing EGFR: p.L858R mutation) cell line were subjected to DNA extraction according to the phenol-chloroform method, and then DNA fragmentation was performed with fragmentation enzyme from NEB Company. Then the DNA of the HCC1975 sample was incorporated into the SK sample at a ratio of 1%. The EGFR:p.L858R mutation frequency was quantified by ddPCR to obtain samples containing the EGFR:p.L858R mutation with known mutation frequency.
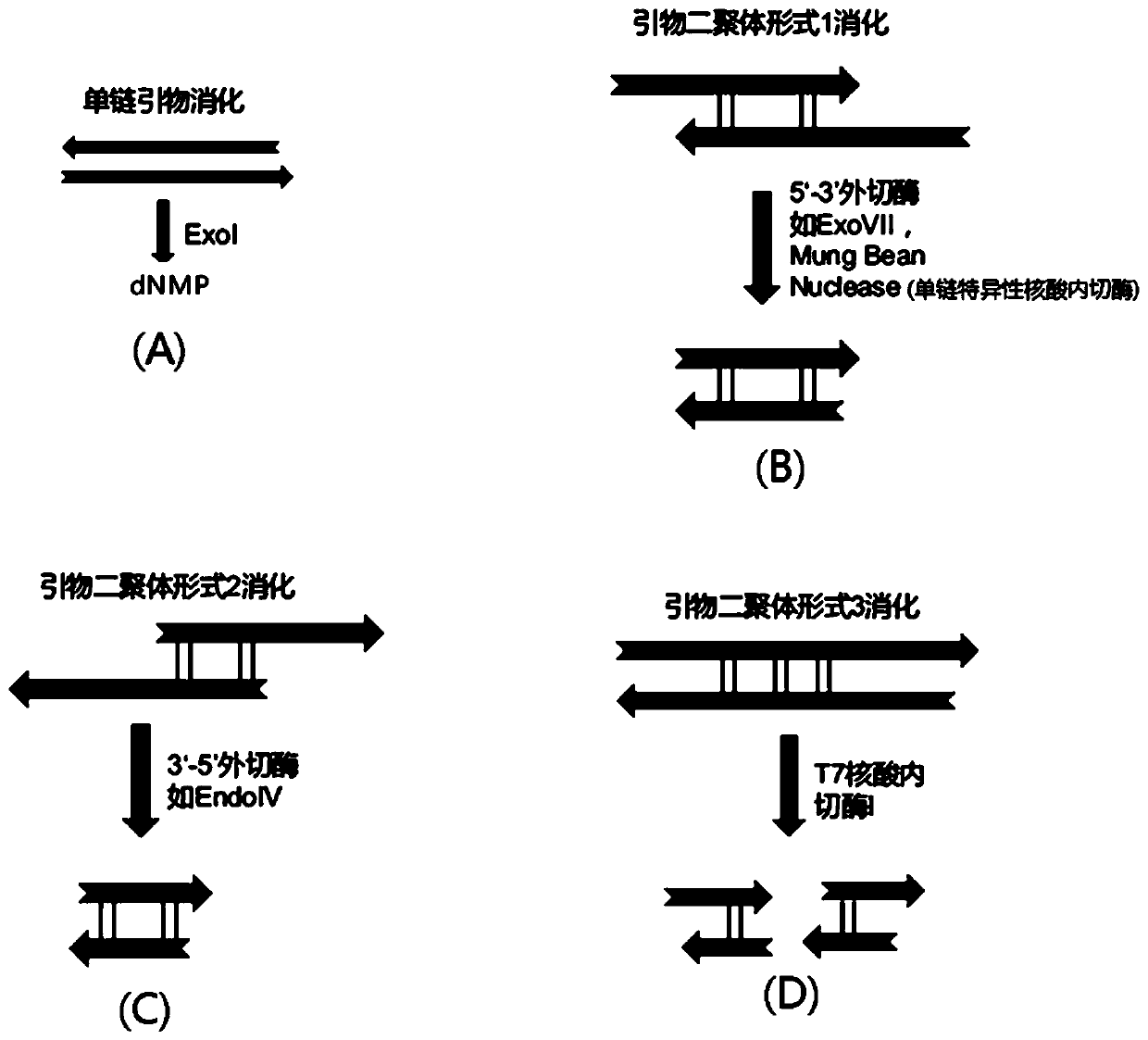
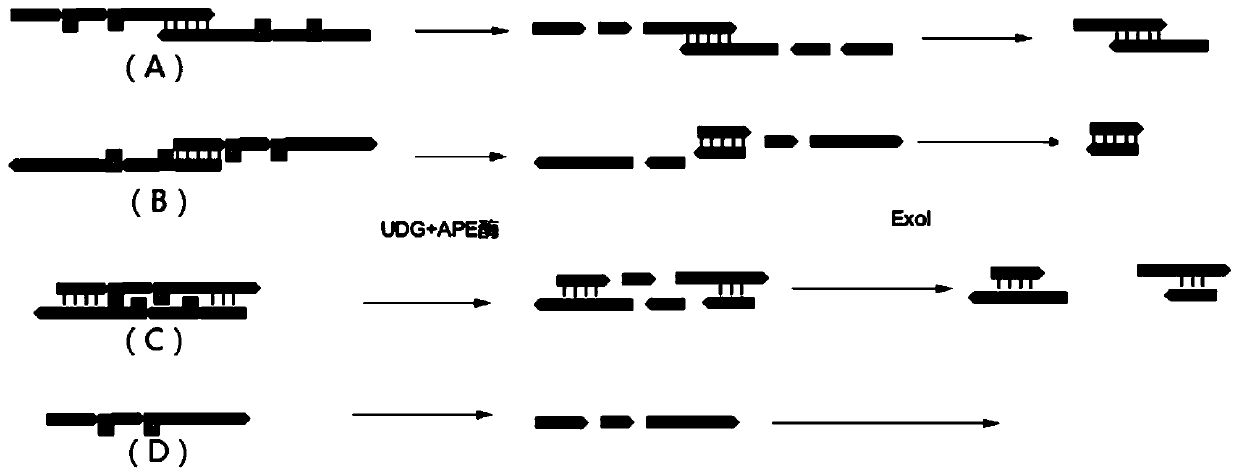
[0100] 2. Primer design. This embodiment designs 2 kinds of PCR primers
[0101] Primer I: PCR primer without U base.
[0102] Primer II: PCR primer with U base.
[0103] The length of the designed primers is 60-70bp, the formed primer-dimer sequence is between 70-100, the length of the designed universal sequencing adap...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com