Reagent kit for detecting enterobacter aerogenes
A technology of Enterobacter aerogenes and kits, which is applied in the field of kits for the detection of Enterobacter aerogenes, can solve the problems of high cost, delay in disease treatment, lack of specific primers, etc., and achieve the effect of low equipment requirements and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0014] Example 1: Sequence Alignment of the rpoB Gene of Enterobacter Bacteria
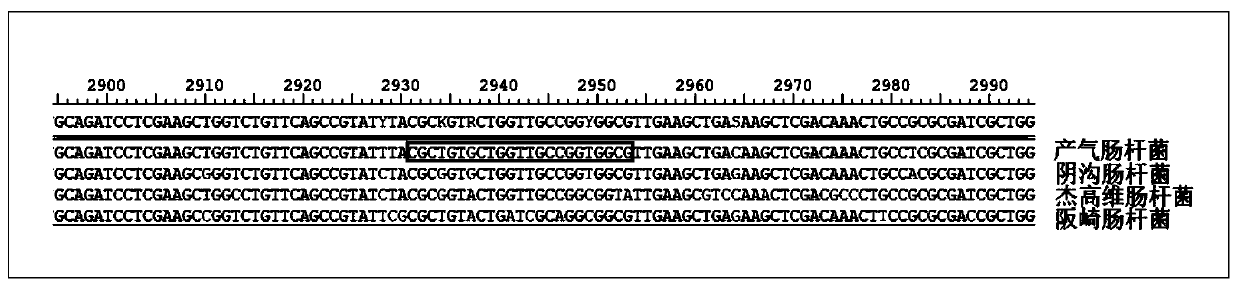
[0015] Such as figure 1 As shown, in the rpoB gene sequence of Enterobacter aerogenes, the 5' end of the guide RNA sequence (selected part) of the present invention is the PAM structure of TTTN. The guide RNA sequence does not have a completely consistent sequence in Enterobacter cloacae, Enterobacter Jagovii, and Enterobacter sakazakii, and there are several base differences. Moreover, there is no PAM structure of TTTN at the 5' end of the homologous sequences in Enterobacter cloacae, Enterobacter jaegavii, and Enterobacter sakazakii. Therefore, using this guide RNA, the Cas12a protein cannot be targeted to Enterobacter cloacae, Enterobacter cloacae, and Enterobacter sakazakii. Enterobacter sakazakii and Enterobacter sakazakii, therefore, only Enterobacter aerogenes can show a positive reaction during the test, and the rest of the Enterobacter genus bacteria can show a negative reaction, which c...
Embodiment 2
[0016] Embodiment 2: The guide RNA specificity determination of the present invention, the steps are as follows:
[0017] (1) Prepare standard strains of Enterobacter aerogenes, Enterobacter cloacae, Enterobacter Jagovius, and Enterobacter sakazakii;
[0018] (2) Take 1ml of the sample to be tested, heat it at 98 degrees Celsius for 5min, and draw 1μl as the test sample;
[0019] (3) Prepare the reaction system, the reaction system is 25 μl, including 1 μl detection sample, 14.75 μl hydrated TwistAmp basickit reaction drying ball (TwistDx company), 0.9 μl 10mM RPA-F (sequence is SEQ No.2) and RPA-R ( Sequence is SEQNo.3), 0.375 μl Ribonuclease Inhibitor (Takara company), 3.5 μl buffer2.1 (NEB company), 1000nM guide RNA (sequence is SEQ No.1), 250nM Cas12a (NEB company), 200nM single-stranded DNA probe Needle (Shanghai Sangong), add water to make up to 25μl;
[0020] (4) React at a constant temperature of 37 degrees Celsius for 30 minutes;
[0021] (5) After the reaction, pl...
Embodiment 3
[0023] Embodiment 3: the detection method based on guide RNA described in the present invention and traditional PCR method specificity and experimental time-consuming comparison
[0024] 1. based on the detection method of guide RNA described in the present invention, the steps are as follows:
[0025] (1) Prepare standard strains of Enterobacter aerogenes, Enterobacter cloacae, Enterobacter Jagovius, and Enterobacter sakazakii;
[0026] (2) Take 1ml of the sample to be tested, heat it at 98 degrees Celsius for 5min, and draw 1μl as the test sample;
[0027] (3) Prepare the reaction system, the reaction system is 25 μl, including 1 μl detection sample, 14.75 μl hydrated TwistAmp basickit reaction drying ball (TwistDx company), 0.9 μl 10mM RPA-F (sequence is SEQ No.2) and RPA-R ( Sequence is SEQNo.3), 0.375 μl Ribonuclease Inhibitor (Takara company), 3.5 μl buffer2.1 (NEB company), 1000nM guide RNA (sequence is SEQ No.1), 250nM Cas12a (NEB company), 200nM single-stranded DNA p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com