DNA barcode primer pair for amplification of liriodendron chinense(Hemsl.)Sarg. and identification method of liriodendron chinense(Hemsl.)Sarg. and germplasm source of liriodendron chinense(Hemsl.)Sarg.
A technology of barcode primers and amplification primers, which is applied in biochemical equipment and methods, recombinant DNA technology, DNA/RNA fragments, etc., can solve the problems of insufficient identification dimension and large variation of morphological markers in Liriodendron tulipifera, and achieve High accuracy and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Liriodendron tulipifera and Liriodendron tulipifera were collected covering the entire distribution area of Liriodendron, a total of 120 samples (sample number 1-120). See Table 1 for sample sources and other relevant information.
[0044] These samples were identified using the DNA barcode primer pair and method of the present invention.
[0045] details as follows:
[0046] 1) Using the improved CTAB method to extract the whole genome DNA of young leaves of the sample to be tested.
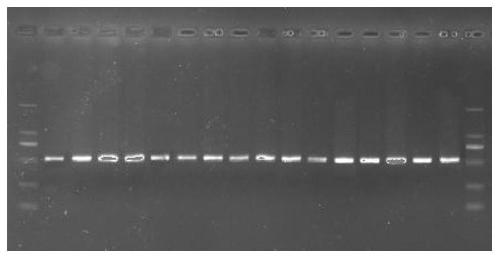
[0047] 2) The obtained whole genome DNA is used as a template, and PCR amplification is performed using the DNA barcode primer pair to obtain an amplified product. The PCR amplification system is calculated in 20 μL, including 10 μL of 2×Taq PCR Master Mix, 2.0 μL of 20-40 ng / L whole genome DNA, 0.6 μL of 10 μmol / L upstream amplification primer, 0.6 μL of 10 μmol / L downstream amplification primer, ddH 2 O 6.8 μL; PCR amplification program includes: 95°C pre-denaturation for 4 minutes...
Embodiment 2
[0052] 153 individuals (sample numbers 13-164) from 43 provenances in the entire distribution area of Liriodendron were collected, covering the largest distribution area of Liriodendron tulipifera. See Table 1 for the source of samples and other related information. Young leaves of all individuals were collected and dried with silica gel for preservation.
[0053] 1) The modified CTAB method was used to extract the total DNA of Liriodendron tulipifera.
[0054] 2) The obtained whole genome DNA is used as a template, and PCR amplification is performed using the DNA barcode primer pair to obtain an amplified product. The PCR amplification system is calculated in 20 μL, including 10 μL of 2×Taq PCR Master Mix, 2.0 μL of 20-40 ng / L whole genome DNA, 0.6 μL of 10 μmol / L upstream amplification primer, 0.6 μL of 10 μmol / L downstream amplification primer, ddH 2 O 6.8 μL; PCR amplification program includes: 95°C pre-denaturation for 4 minutes; 95°C denaturation for 40 s, 59°C anne...
Embodiment 3
[0059] Sample source and operating steps are the same as in Example 2.
[0060] According to the results of sequencing and sequence comparison, it was found that starting from 375bp, samples No. 13 to 26 had 8 A base repeats, which was consistent with the standard sequence SEQ ID No.6 of the tropical provenance of Liriodendron tulipifera of the present invention, and was identified as a tropical Provenance, the remaining samples are all 9 A base repeats starting from 375bp. For the sequence alignment of samples numbered 24-26 and 100-102 with DNA barcode sequences (SEQ ID No.5 and SEQ ID No.6) in the 320-395bp region, see Figure 4 According to the source of the control samples, it was found that samples 13-26 all came from tropical regions, and the rest of the samples all came from subtropical regions. Therefore, the identification accuracy rate of the barcode of the present invention is 100%.
[0061] It can be known from the above examples that the method for identifying ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com