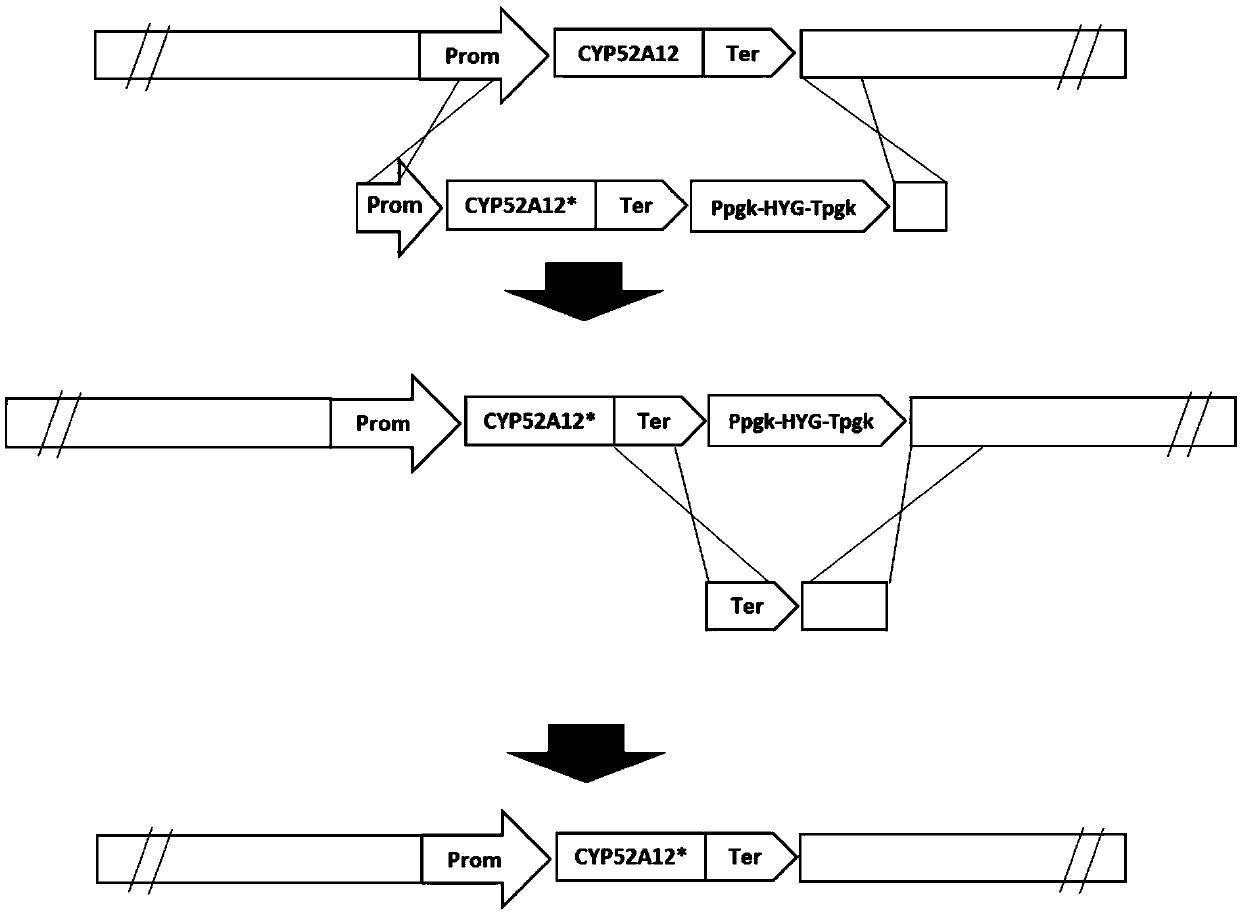
Directed evolution of CYP52A12 gene and application of gene in production of binaryacid
A CYP52A12, dibasic acid technology, applied in application, genetic engineering, plant genetic improvement, etc., to achieve the effect of shortening the fermentation cycle, significant cost advantages, and high concentration of acid production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0089] Embodiment 1 culture medium, culture fermentation method and dibasic acid detection method
[0090] 1. YPD medium, the formula (w / v) is: 2% peptone, 2% glucose and 1% yeast extract (OXOID, LP0021). 1.5-2% agar powder needs to be added to the solid medium.
[0091] During cultivation, a single colony can be placed in a 2ml centrifuge tube containing 1ml YPD liquid medium, and cultured on a shaker at 250RPM for 1 day at 30°C.
[0092] 2, seed medium, formula (w / v) is: sucrose 10~20g / L, yeast extract 3~8g / L, corn steep liquor (abbreviated as corn steep liquor, total nitrogen content 2.5wt%) 2~4g / L for industrial fermentation L, KH 2 PO 4 4-12g / L, 0.5-4g / L urea (sterilized separately at 115°C for 20min), and the fermentation substrate is n-dodecane 20mL / L.
[0093] When cultivating, insert the bacterial solution cultivated in step 1 into a 500mL shake flask containing 30mL seed medium, the inoculum amount is 3-5%, and cultivate to OD at 250rpm and 30°C on a shaker 620...
Embodiment 2
[0098] Example 2 Preparation of CYP52A12 Mutation Template
[0099] 1. Preparation of CYP52A12 mutation template.
[0100] Genomic DNA of Candida CCTCC M 2011192 was extracted using the Ezup Yeast Genomic DNA Rapid Extraction Kit (Sangon, Cat. No. 518257). In order to improve the efficiency of cell wall crushing, liquid nitrogen grinding was used to crush the cell wall. Error-prone PCR was performed using the genomic DNA obtained by this method as a template.
[0101] 2. Error-prone PCR
[0102] Adjust Mg 2+ Concentration (2-8mM), using Taq DNA polymerase (Takara, Cat. No. R001B) for error-prone PCR amplification of CYP52A12 gene. (PCR conditions are: step 1: 98°C for 30s, step 2: 98°C for 10s, 55°C for 30s, 72°C for 2m for 20s, a total of 35 cycles, step 3: 72°C for 5m), the primers are as follows:
[0103] CYP52A12-F: 5'-CAAAACAGCACTCCGCTTGT-3' (SEQ ID NO: 1),
[0104] CYP52A12-R: 5'-GGATGACGTGTGTGGCTTGA-3' (SEQ ID NO: 2),
[0105] After the PCR product was subjected ...
Embodiment 3
[0106] Example 3 Preparation of Homologous Recombination Template
[0107] All DNA fragments used in this example HS high-fidelity DNA polymerase (Takara, R040A) amplified. After 1% agarose gel electrophoresis, the purified DNA fragments were recovered with the Axygen gel recovery kit.
[0108] (1) Amplification of the resistance selection marker (HYG, hygromycin resistance gene), the amplification template is the vector pCIB2 (SEQ ID NO: 3) owned by our company, and the primer sequence is as follows:
[0109] CYP52A12_HYG-F:
[0110] 5'-TCAAGCCACACACGTCATCCGCATGCGAACCCGAAAATGG-3' (SEQ ID NO: 4),
[0111] CYP52A12_HYG-R:
[0112] 5'-GATGTGGTGATGGGTGGGCTGCTAGCAGCTGGATTTCACT-3' (SEQ ID NO: 5).
[0113] The PCR reaction conditions are as follows:
[0114] Step 1: 98°C for 30s,
[0115] Step 2: 98°C for 10s, 55°C for 30s, 72°C for 1m for 50s, 5 cycles,
[0116] Step 3: 98°C for 10s, 72°C for 2m, 25 cycles,
[0117] Step 4: 72°C for 5m.
[0118] The obtained product is c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com