Method for screening gene keywords from PubMed literatures
A technology for screening genes and keywords, applied in the field of bioinformatics, can solve problems such as low accuracy rate and high false positive rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
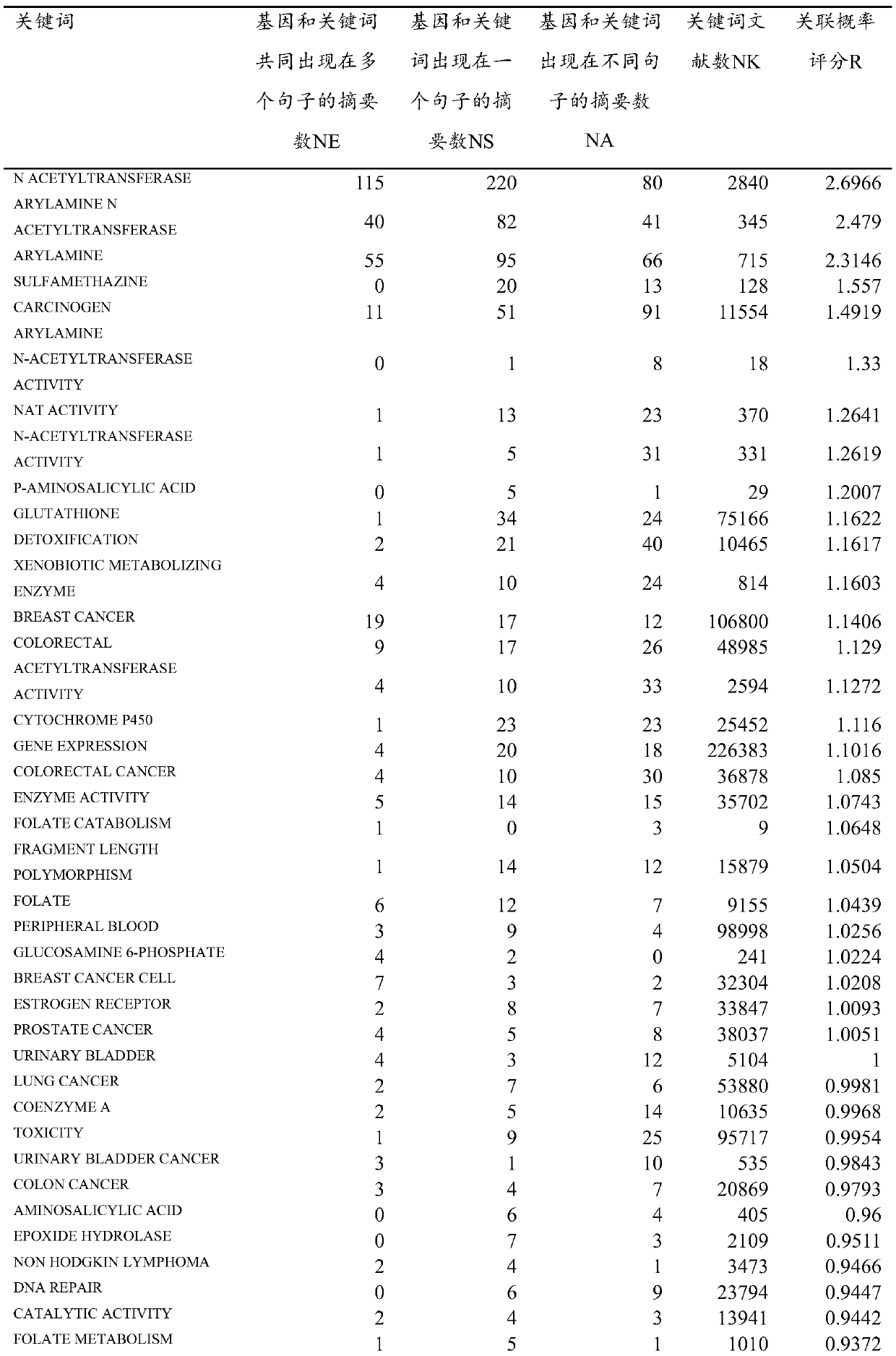
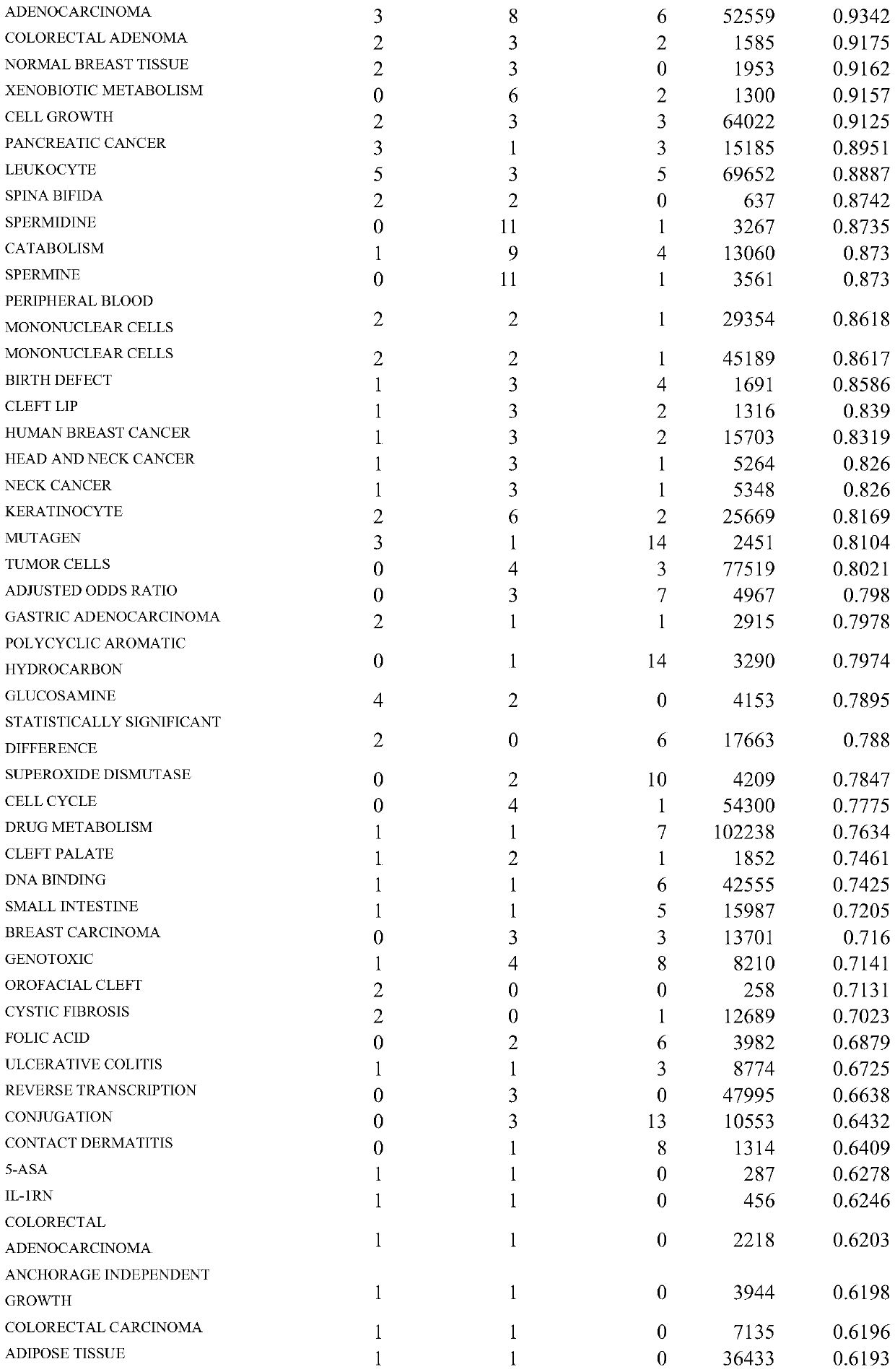
[0043]1. Obtain PubMed's annual update literature and daily update literature through MEDLINE / PubMed FTP (ftp: / / ftp.ncbi.nlm.nih.gov / pubmed / ), and extract the relevant information of the NAT1 gene from the downloaded XML file, such as PMID (PubMed ID), title, abstract information, localize the PubMed literature related to the NAT1 gene, identify the NAT1 gene name in the PubMed abstract, and correspond to the correct Entrez Gene ID (GID), determine the NAT1 gene related abstract, use Perl's Text::Sentence module segments NAT1 gene-related summaries into sentences (SIDs) and identifies NAT1 gene-related sentences. Finally, a total of 583 gene-related abstracts and 1862 gene-related sentences related to the NAT1 gene were found.
[0044] 2. Use the search engine Sphinx (http: / / sphinxsearch.com / ) to perform full-text indexing of NAT1 gene-related documents. Since Sphinx itself cannot store text fields, we combine it with MySQL. The MySQL database stores documents and their corres...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com