Single-cell ATAC-seq data analysis method
A technology of data analysis and analysis methods, applied in the direction of sequence analysis, bioinformatics, informatics, etc., to achieve the effect of displaying various forms, increasing readability, and clear layers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
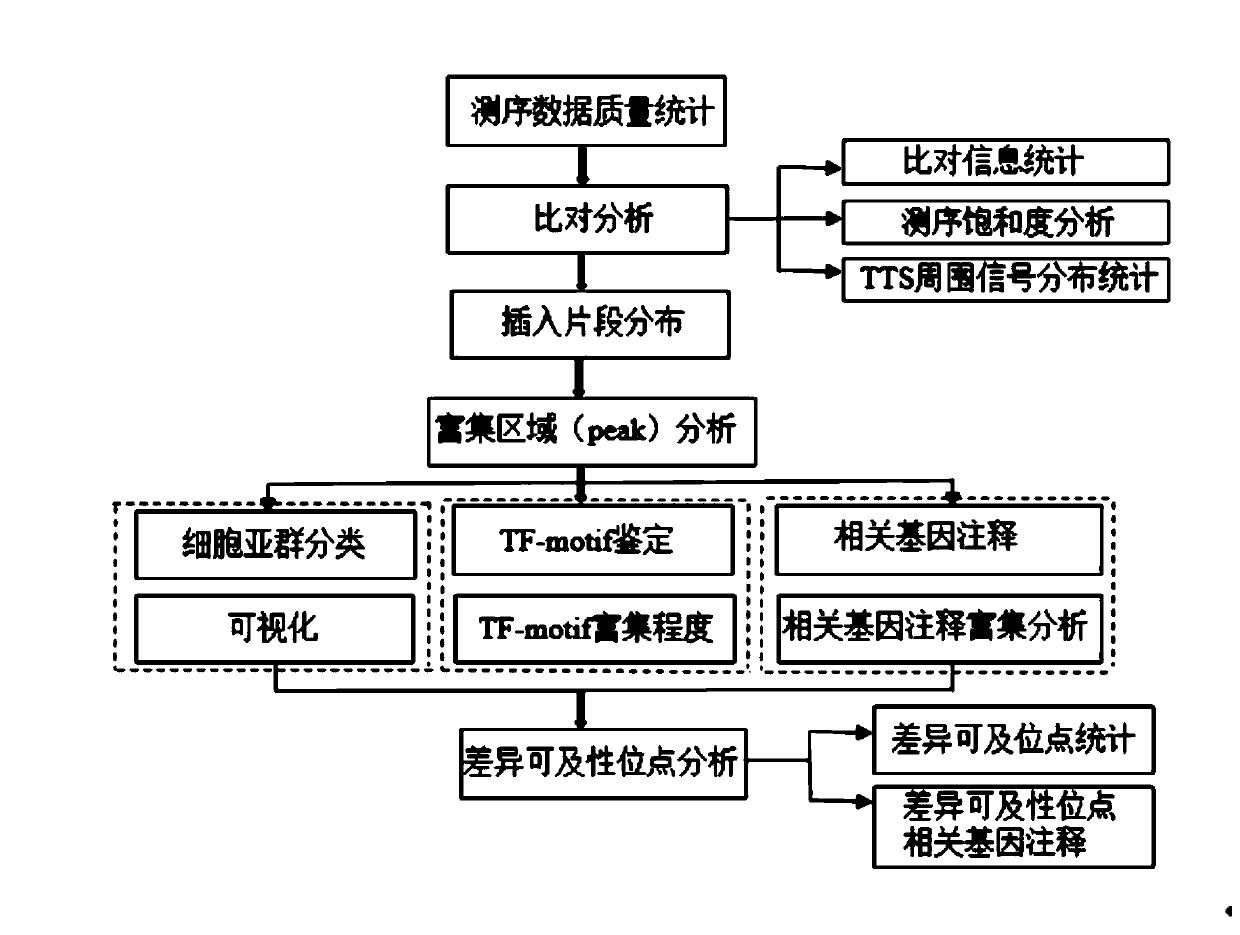
[0029] The single-cell ATAC-seq data analysis process in this embodiment includes the following steps:
[0030] Step S1: Perform data analysis and quality control on the raw sequencing data to obtain high-quality data for subsequent analysis. Mainly use the Cell Ranger software to filter and correct the wrong barcodes in the sequencing raw data. Compare each barcodes sequence with the known barcodes sequence in the database, find the barcodes with a base mismatch of ≤ 2bp with the known barcodes, and score according to the abundance of the read barcodes and the quality value of the mismatched bases. Barcodes with values greater than 90% are considered correct barcodes.
[0031] Step S2: Alignment analysis, use cutadapt to identify the reverse complementary sequence of the primer at the end of the reads, and remove it from the reads sequence, then use BWA-MEM to align the trimmed reads to the reference genome, and then use Duplication analysis to determine the unique alignme...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com