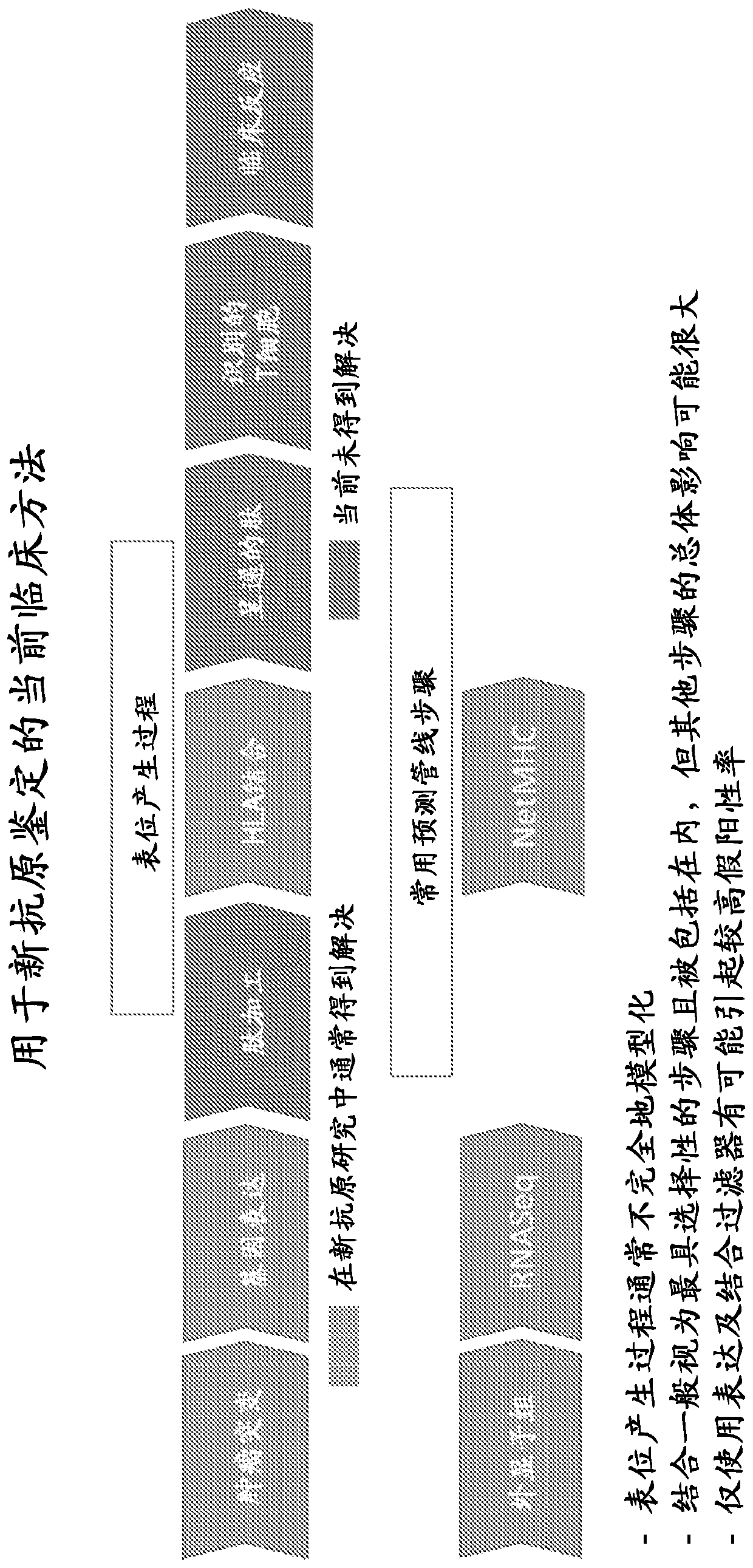
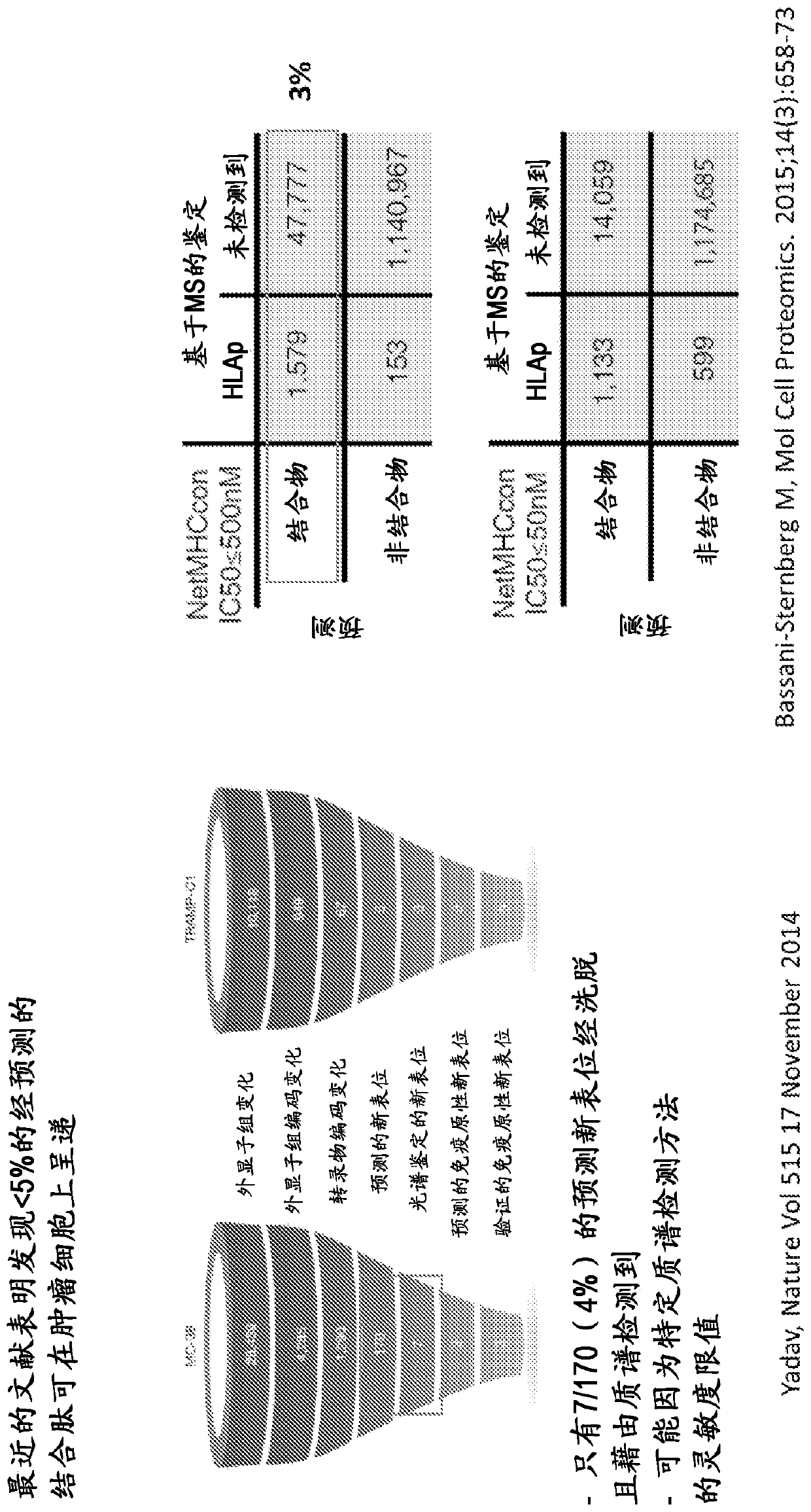
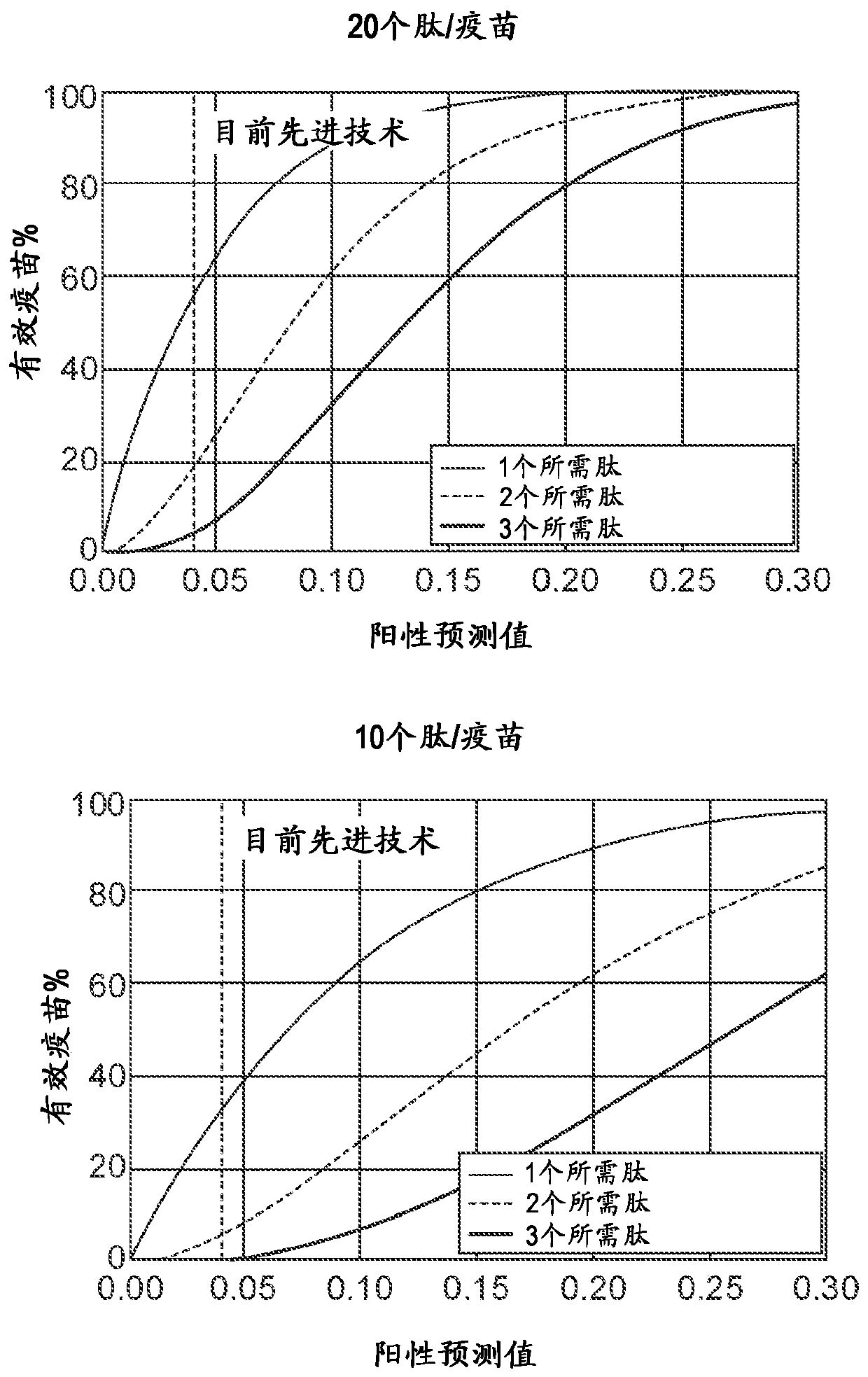
Viral delivery of neoantigens
An antigen, adenovirus technology, applied in the direction of viruses, cancer antigen components, viruses/phages, etc., can solve the problems of missing candidate neoantigen somatic mutations, inefficient use of vaccines or autoimmune risks.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0657] X.C.1. Example 1: Maximum value of independent allele model
[0658] In one embodiment, the training module 316 makes the peptide p associated with a set of multiple alleles H k Estimated probability of presentation u k With the presentation probability u of each MHC allele h in the set H determined based on cells expressing a single allele k h∈H The change in is modeled as described above in conjunction with equations (2)-(11). Specifically, the possibility of presentation u k Can be u k h ∈ H Any function. In one embodiment, as shown in equation (12), the function is the maximum function, and the probability u k It can be determined as the maximum presentation probability of each MHC allele h in the set H.
[0659]
[0660] X.C.2. Example 2.1: Function-of-Sums model of sum
[0661] In one embodiment, the training module 316 makes the peptide p k Estimated probability of presentation u k Modeling:
[0662]
[0663] Where element a h k For peptide sequence p k Multiple assoc...
example 22
[0673] X.C.3. Example 2.2: A functional model using the sum of allelic non-interacting variables In one embodiment, the training module 316 incorporates the allelic non-interacting variables and makes the peptide p by the following formula k Estimated probability of presentation u k Modeling:
[0674]
[0675] Where w k Indicates the encoded related peptide p k The allelic non-interacting variable. Specifically, the parameter set θ of each MHC allele h h And related allelic non-interacting variable parameter set θ w The value of can be determined by h And θ w The loss function of i is minimized to determine each instance of the subset S of the training data 170 generated by cells expressing a single MHC allele and / or cells expressing multiple MHC alleles. Correlation function g w It can be the correlation function g introduced in section X.B.3 above w In any form.
[0676] Therefore, according to equation (14), the peptide sequence p k The probability of presentation that will be p...
example 31
[0687] X.C.4. Example 3.1: Model using implicit independent allele likelihood
[0688] In another embodiment, the training module 316 makes the peptide p k Estimated probability of presentation u k Modeling:
[0689]
[0690] Where element a h k For peptide sequence p k The associated multiple MHC alleles h∈H are 1, Is the implicit independent allele presentation possibility of MHC allele h, vector v is element v h Corresponds to The vector of s(·) is a function of the mapping element v, and r(·) is a clipping function, which reduces the input value to a given range. As described in more detail below, s(·) may be a summation function or a second-order function, but it should be understood that in other embodiments, s(·) may be any function, such as a maximum function. The value of the parameter set θ related to the possibility of implicit independent alleles can be determined by minimizing the loss function related to θ, where i is determined by cells expressing a single MHC alle...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com