Application of Notch1, p16<INK4a> and hTERC genes as screening markers for cervical cancer
A p16ink4a, cervical cancer technology, applied in the field of clinical medicine, can solve the problems of excessive cell proliferation, insignificant correlation between the occurrence of cervical cancer, and low reliability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] 1. Materials and methods
[0027] 1.1 Paraffin section of cervical lesion tissue and positive determination of HPV16
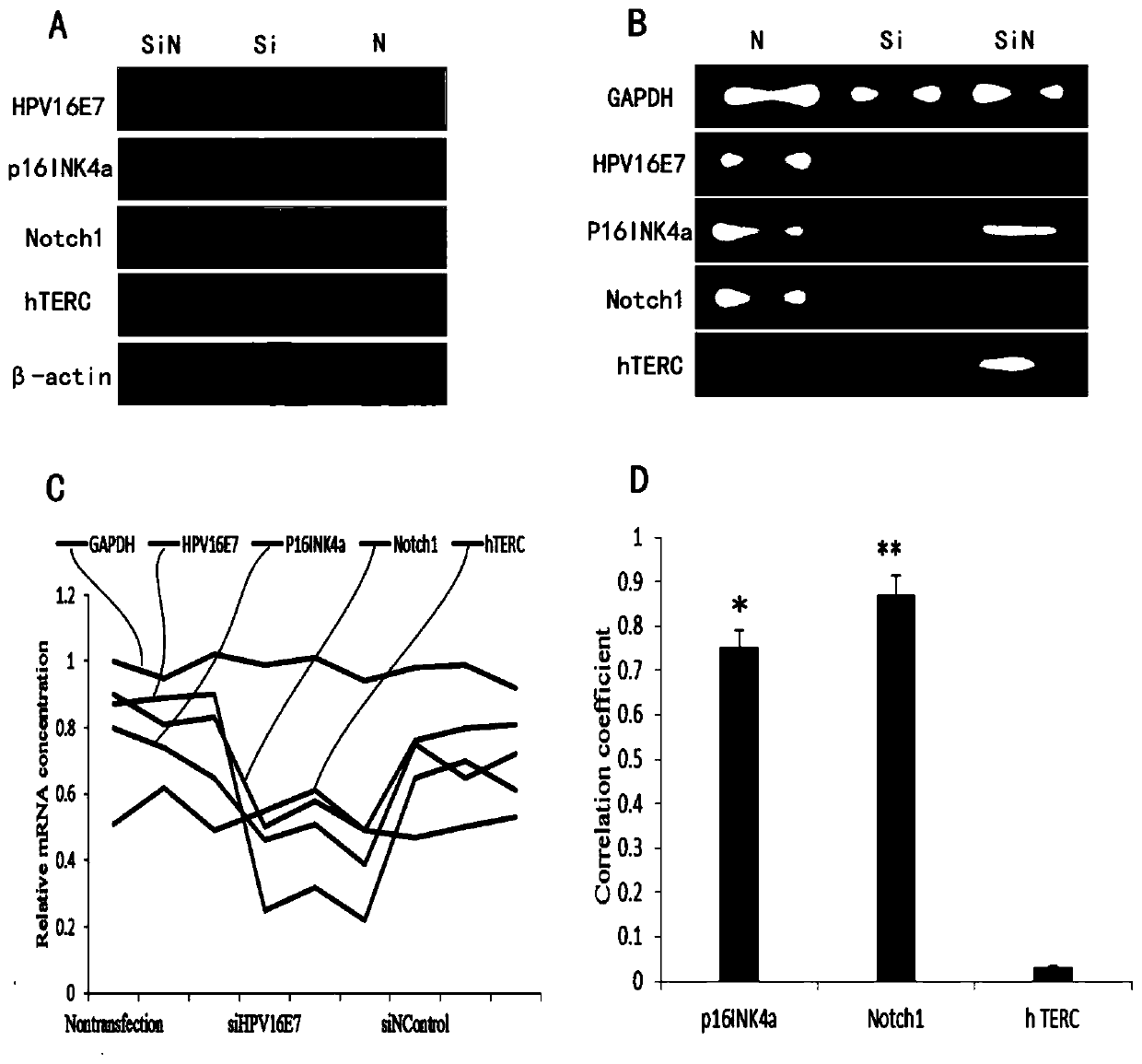
[0028] From September 2016 to September 2017, patients with cervical diseases who visited the Affiliated Hospital of Inner Mongolia University for Nationalities and underwent pathological examination were selected, and 23 kinds of HPV genotyping detection kits (PCR fluorescent detection kits) provided by Guangzhou Kaipu Biotechnology Co., Ltd. were used. Acupuncture method), strictly follow the instruction manual to extract HPV DNA and perform PCR amplification, and use ABIQuantistudio DX fluorescence quantitative PCR instrument (ABI company, the United States) for HPV genotyping detection, and 226 patients who were positive for HPV16 were detected, of which 45 Cervical cancer, 35 cases of CIN-Ⅲ, 32 cases of CIN-Ⅱ, 38 cases of CIN-Ⅰ, 42 cases of uterine leiomyoma, 34 cases of chronic cervicitis. The age range was 20 to 71 years old, with an average age...
Embodiment 2
[0043] 2. Results
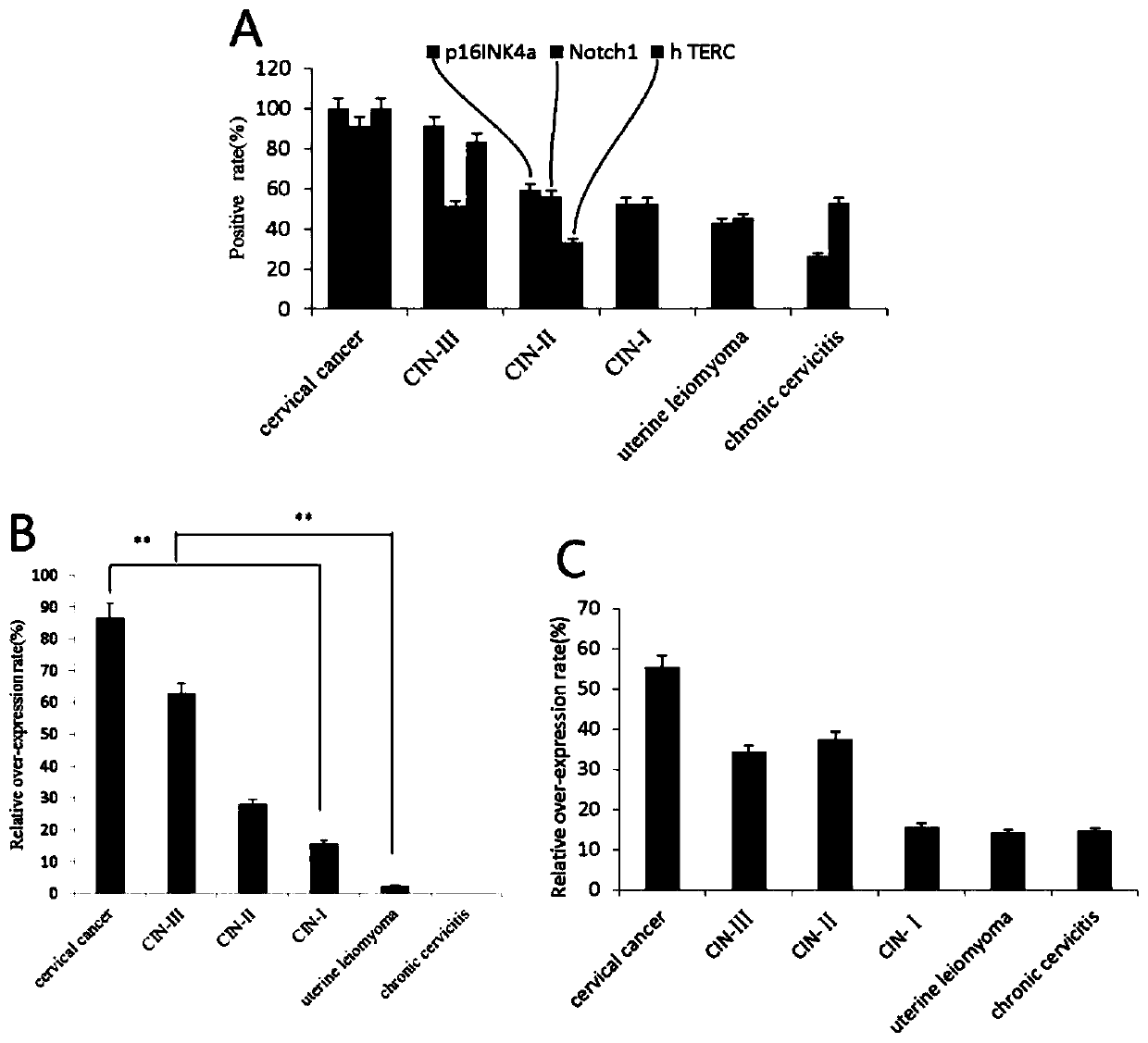
[0044] 2.1 p16 INK4a , Notch1 and hTERC There were significant differences in the positive expression of the gene in each HPV16 positive cervical lesion tissue.
[0045] The detection of HPV16-positive cervical lesion tissue by immunohistochemistry and fluorescence in situ hybridization showed that see Figure 4 , in cervical cancer, CIN-III, CIN-II, CIN-I, uterine leiomyoma, and chronic cervicitis, p16 INK4a The positive expression rates of the proteins were 100.00% (45 / 45), 91.43% (32 / 35), 59.38% (19 / 32), 52.63% (20 / 38), 42.86% (18 / 42), 26.47% ( 9 / 34); the positive expression rates of Notch1 protein were 91.11% (41 / 45), 51.43% (18 / 35), 56.25% (18 / 32), 52.63% (20 / 38), 45.23% (19 / 34 42), 52.94% (18 / 34); hTERC The positive amplification rates of the genes were 100.00% (25 / 25), 83.33% (10 / 12), 33.33% (4 / 12), 0% (0 / 12), 0% (0 / 9), and 0% (0 / 30), the differences were significant, and p16INK4a was the most significant. And in the detection of the same...
Embodiment 3
[0047] 2. Results
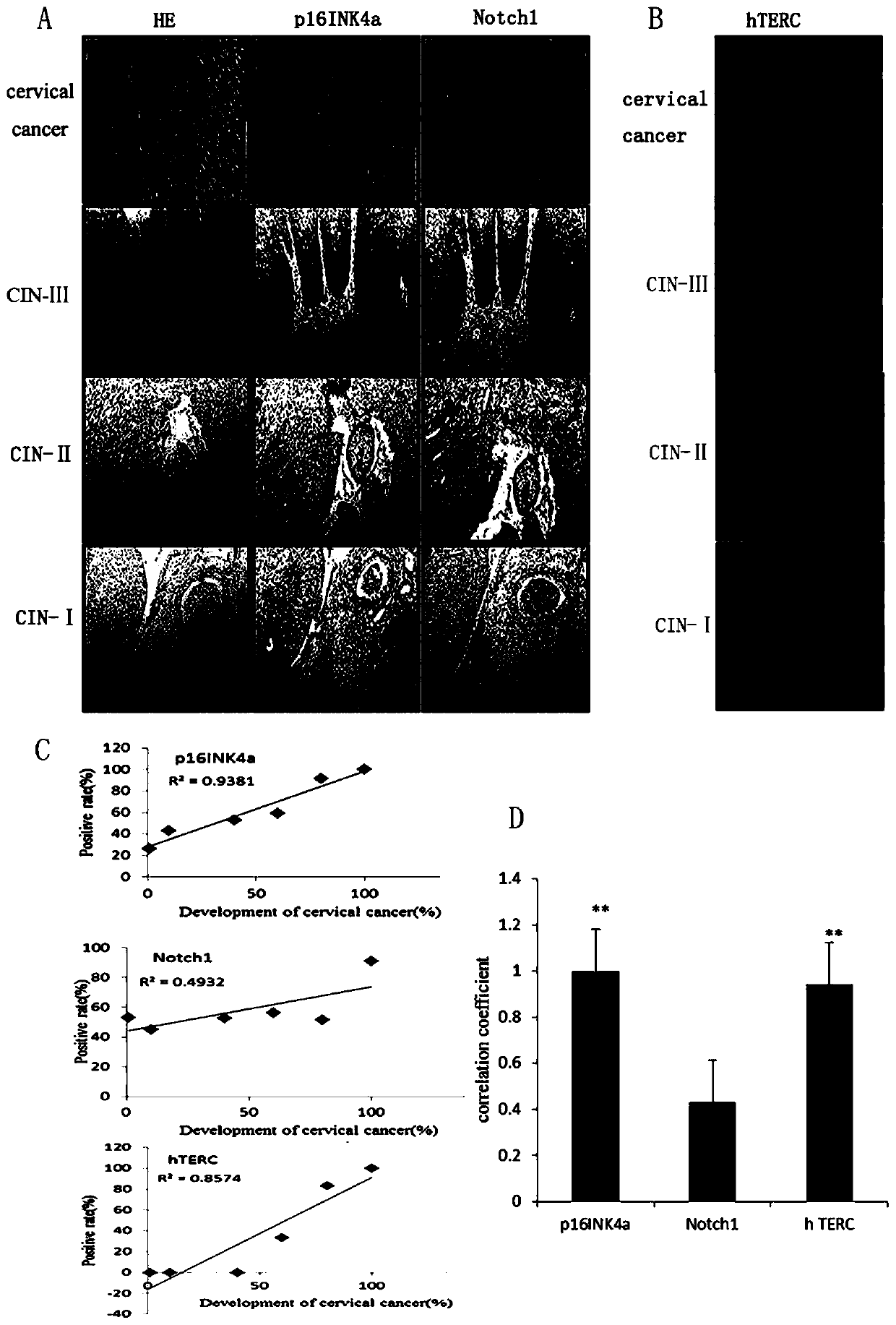
[0048] 2.2 p16 INK4a Gene most strongly associated with development and progression of cervical cancer
[0049] p16 INK4a , Notch1 and hTERC The expression of the gene is constantly changing with the occurrence and development of cervical cancer, showing a positive correlation. p16 INK4a , Notch1 In immunohistochemistry, the gene obviously showed an increase in the number of stained cells and the degree of staining in the tissue ( figure 2 A), hTERC gene in fluorescence in situ hybridization showed that the number of cells with gene amplification and the copy number of the gene increased ( figure 2 B). exist figure 2 In A we can also find p16 INK4a It shows high specificity and sensitivity in the occurrence and development of cervical cancer. Compared with HE staining, its expression only occurs in the lesion and its expression degree is distinct in each stage of cervical cancer development. and Notch1 The expression of the gene is not...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com