RNA analyzing method of paraffine section tissue
A technology of paraffin sectioning and analysis methods, applied in sequence analysis, biochemical equipment and methods, instruments, etc., can solve the problems of low quality of sequencing data comparison and technical incompatibility, and achieve accurate and effective evaluation results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
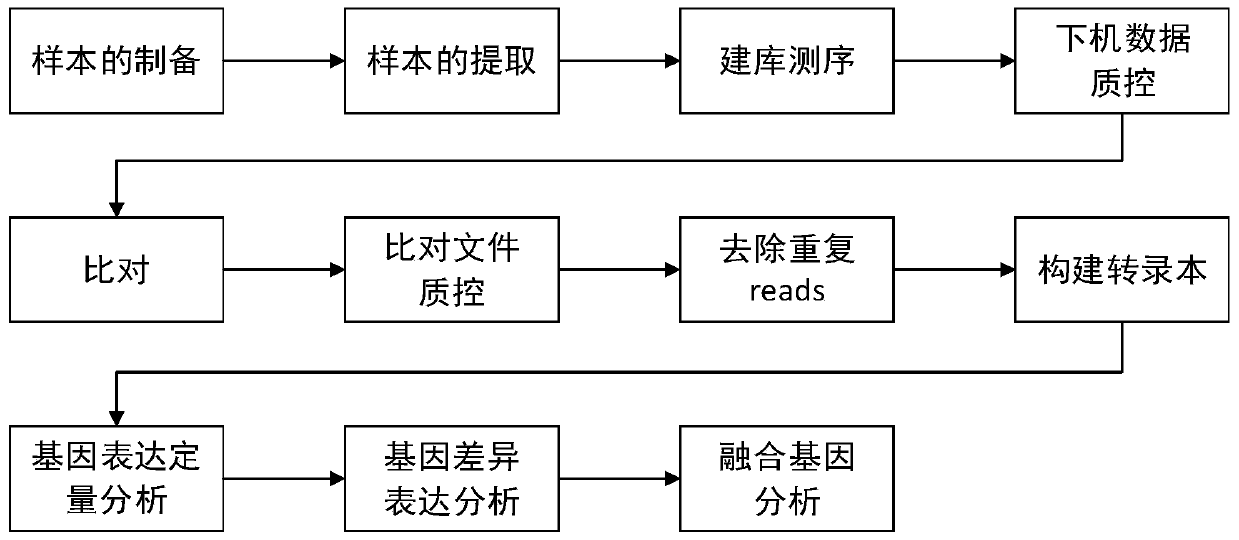
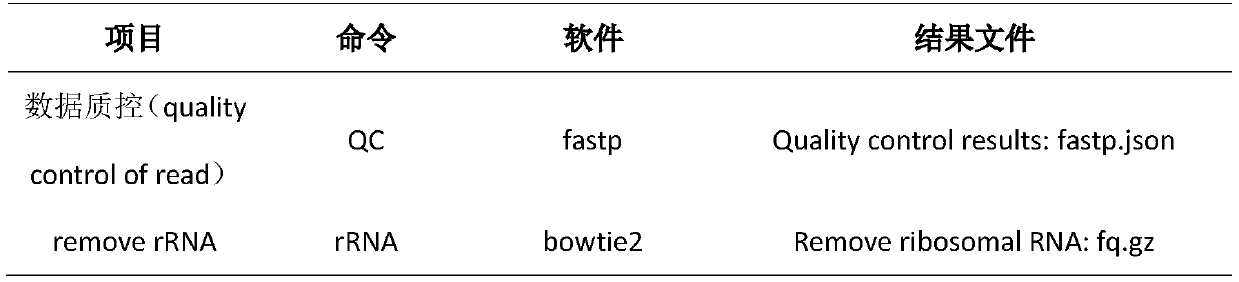
[0041] like figure 1 As shown, a method for RNA analysis of paraffin section tissue includes the following steps: DNA degradation is performed on paraffin section tissue, and sample RNA is extracted; a nucleic acid library of paraffin section samples is prepared, and based on the library, the sample RNA is sequenced; The sample data obtained by sequencing is subjected to quality control, and rRNA data is removed; the sample data after the quality control is compared with the reference genome, and the comparison results are subjected to quality control; the sample data after the quality control is subjected to quality control; Transcriptome assembly and transcript quantification are used to quantitatively analyze gene expression; based on the transcript quantification results, gene differential expression analysis is performed; at the same time, fusion gene analysis can also be performed using the method described in this example. The software for each step and its result files...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com