Method for constructing DNA (deoxyribonucleic acid) methylation library and application of method
A technology of methylation and DNA molecules, applied in chemical libraries, biochemical equipment and methods, combinatorial chemistry, etc., can solve the problem of methylation in other regions such as CHH and CHG regions, which may also be related to tumor occurrence and cannot be detected Detect and other problems to achieve the effect of avoiding template loss, saving time, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
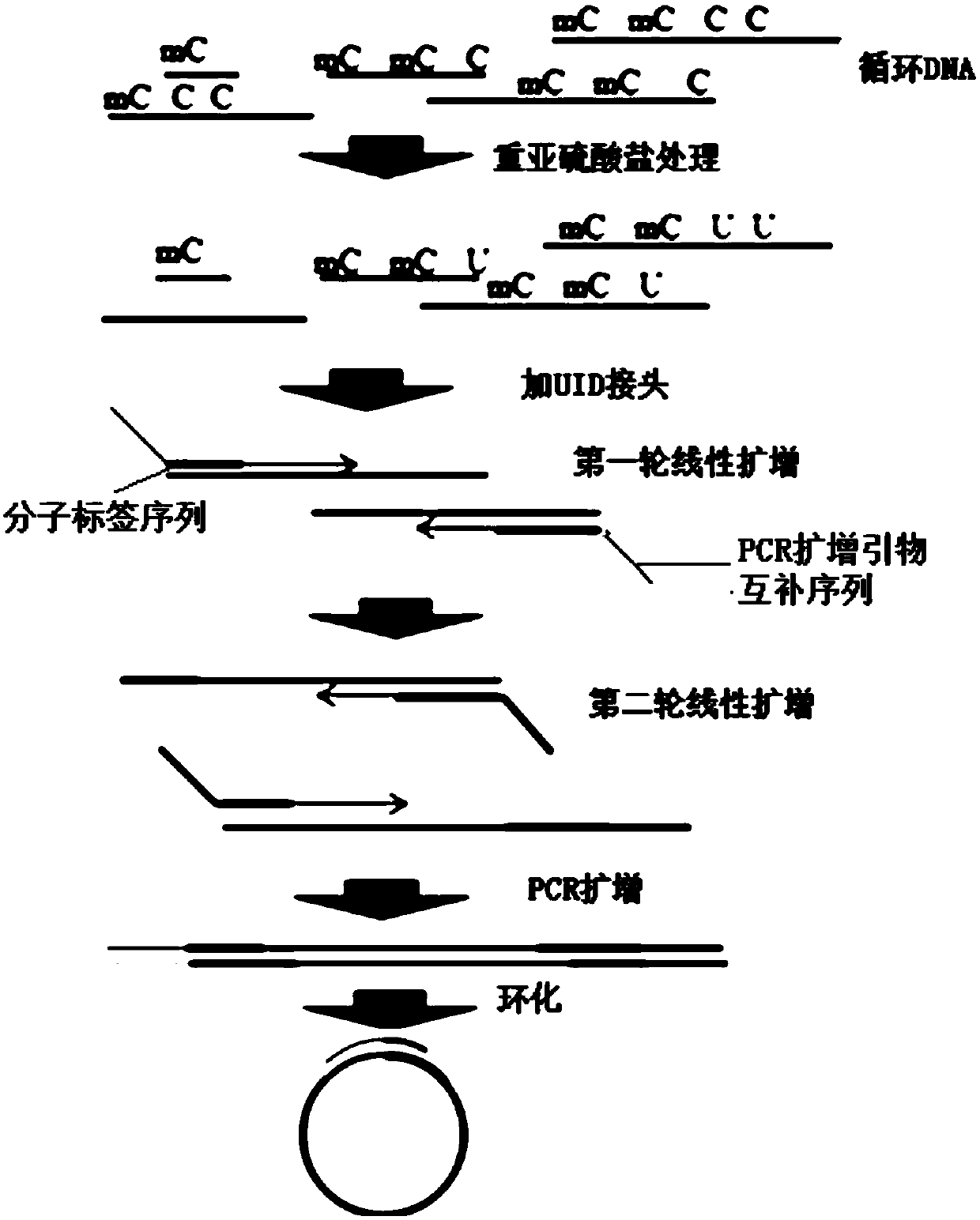
[0050] According to another specific embodiment of the present invention, the method for constructing a DNA methylation library further includes; performing PCR amplification on the DNA molecule connected by the linker sequence, so as to obtain a PCR amplification product; The amplification product is denatured into single-stranded DNA, and the single-stranded DNA is circularized into circular single-stranded DNA. According to an embodiment of the present invention, the amplification product is denatured into a single-stranded DNA by high temperature, and the single-stranded DNA is circularized under the action of a circularization primer, T4DNA Ligase (T4DNA Ligase) and ATP to obtain a circularized product. The circularization product can be subjected to the action of exo I and exo III to obtain a single-stranded circular DNA library that can be sequenced on the machine. Preferably, the PCR amplification product is purified by magnetic beads, and then denatured into a single ...
Embodiment 1
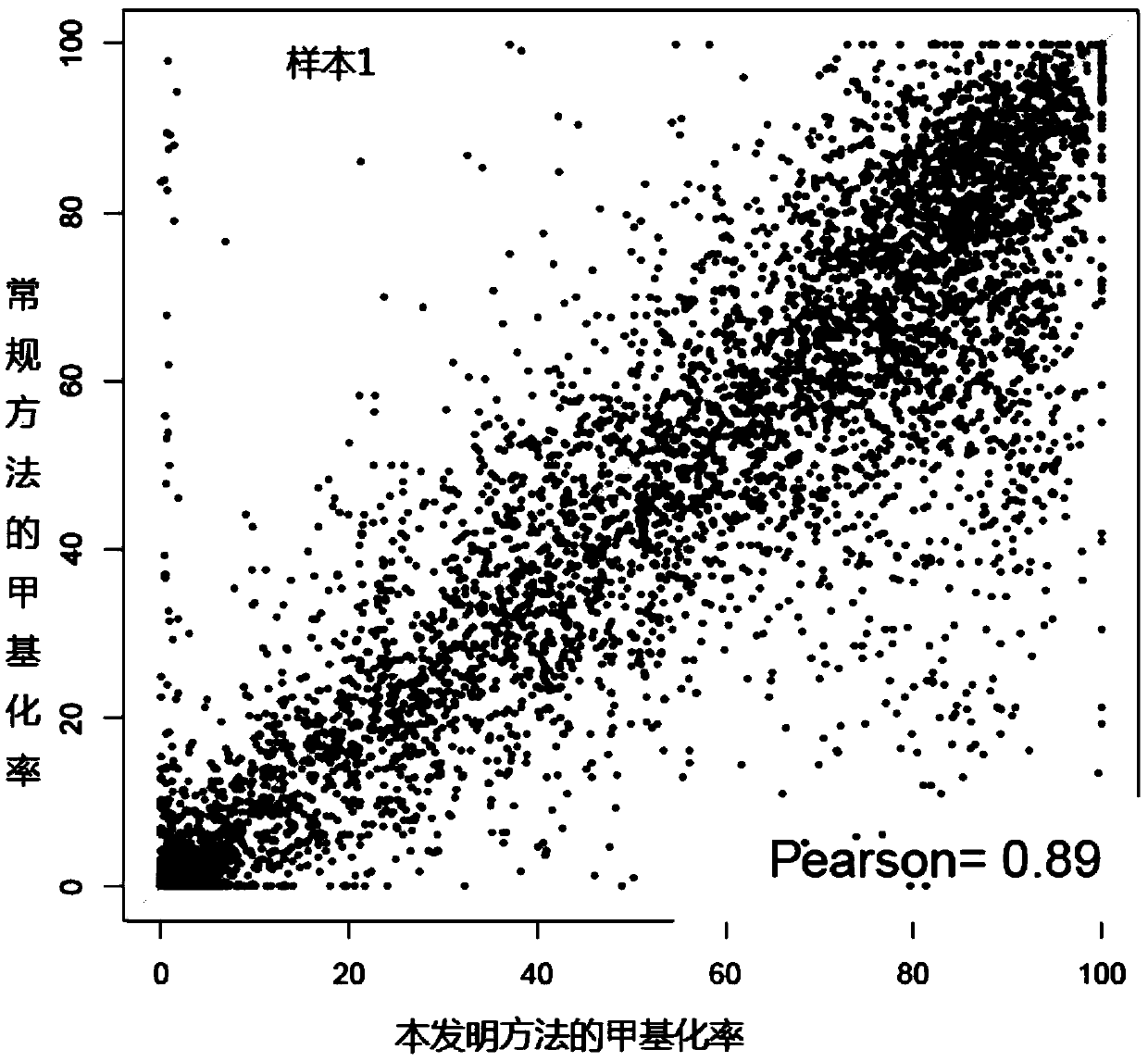
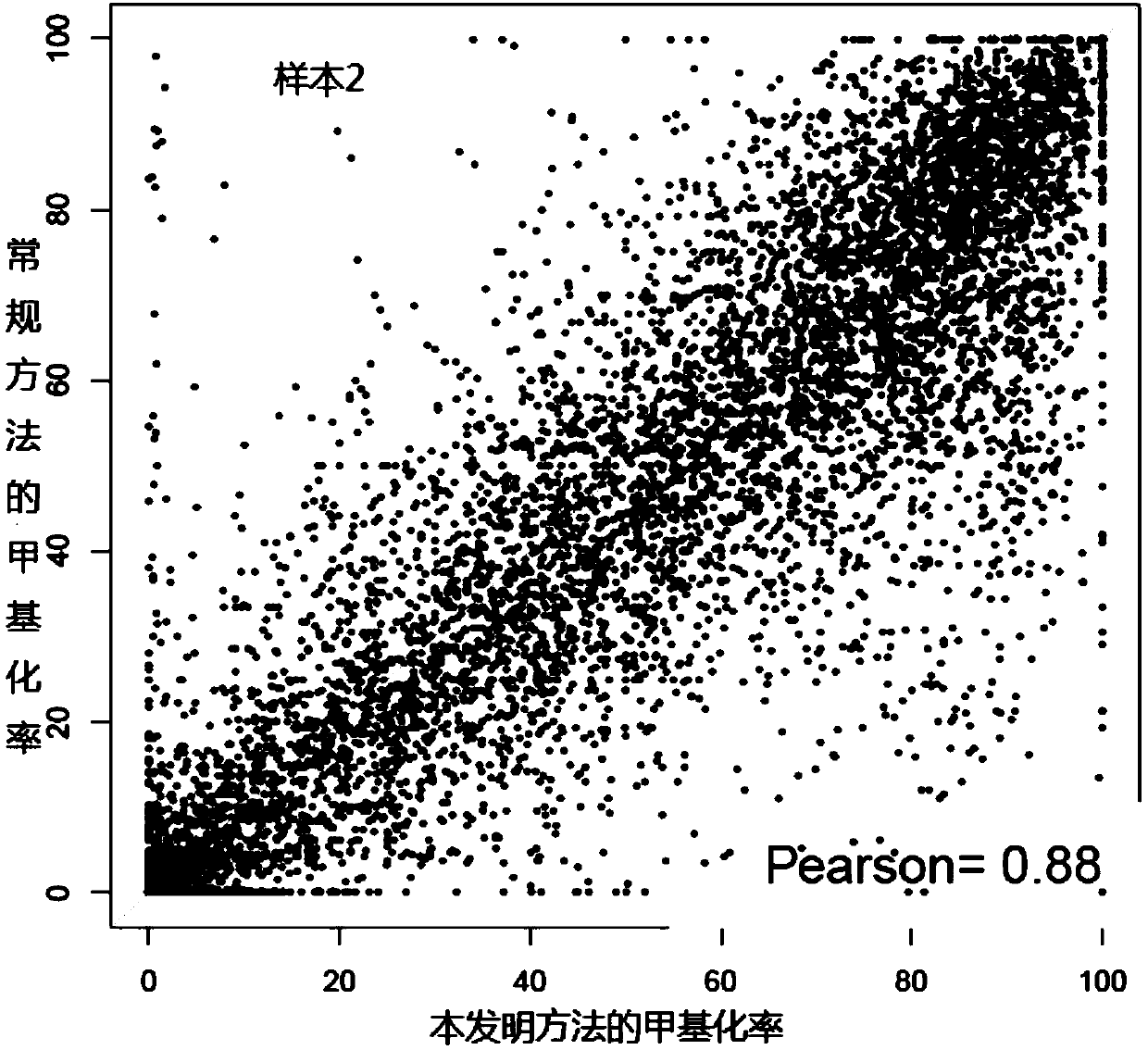
[0058] In this embodiment, circulating tumor DNA is obtained by using the blood samples of tumor patients, and then bisulfite-converted single-stranded DNA molecules are obtained by using bisulfite conversion treatment; and through linear amplification, single-stranded DNA molecules are respectively Linker sequences are connected to both ends of the adapter sequence; PCR amplification is performed using the DNA molecule connected by the adapter sequence to obtain a PCR amplification product; the PCR amplification product is denatured into single-stranded DNA and circularized into circular single-stranded DNA. Use the BGISEQ sequencing platform to obtain the sequencing results, and obtain the DNA methylation detection results of tumor patients.
[0059] At the same time, the detection results were compared with the methylation detection results of the tumor tissues of the above-mentioned tumor patients by using the method of routine whole genome library construction and sequenci...
PUM
| Property | Measurement | Unit |
|---|---|---|
| conversion efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com