Methods and reagents for analyzing protein-protein interfaces
A protein, C2-C9 technology, applied in analytical materials, chemical instruments and methods, biochemical equipment and methods, etc., can solve problems such as insurmountable
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0510] Example 1: Synthesis of certain crosslinkers
[0511] Synthesis of Cyclosporine Analogs Containing Acrylamide
[0512]
[0513] All reagents and solvents were purchased from Sinopharm Chemical Reagent Co.Ltd. Fmoc-amino acid, HATU, HOAT, H-Ala-2-Cl-(Trt) resin (0.36mmol / g), H-Leu-2-Cl-(Trt) resin (0.30mmol / g), H-Phe- 2-Cl-(Trt) resin (0.35 mmol / g) and H-Thr(tBu)-2-Cl-(Trt) resin (0.36 mmol / g) were purchased from GLBiochem (Shanghai) Ltd.
[0514] Coupling of linear peptides was performed on an automated synthesizer using the standard Fmoc SPSS program.
[0515] General Method A: By using TETRAS TM The synthesizer synthesized linear peptides on a scale of 0.025 mmol resin. The general protocol is as follows: 2x NMP, 30 s; 1x 20% (vol / vol) piperidine in NMP, 15 min; 5x NMP, 30 s; amino acid (3 equivalents) solution in NMP was added to the vessel containing the resin, then The solutions of HATU and DIEA in DMF were added respectively, and coupled for 45min; 3x NMP ...
Embodiment 2
[0673] Example 2: Synthesis of certain conjugates
[0674] General Protocol: The protocol describes a method for forming target protein-compound conjugates.
[0675] Reagents: Compound (internal) and mammalian target protein (internal) in 100% DMSO
[0676] Equipment: Micro PROTEAN TGX gel (Bio-Rad)
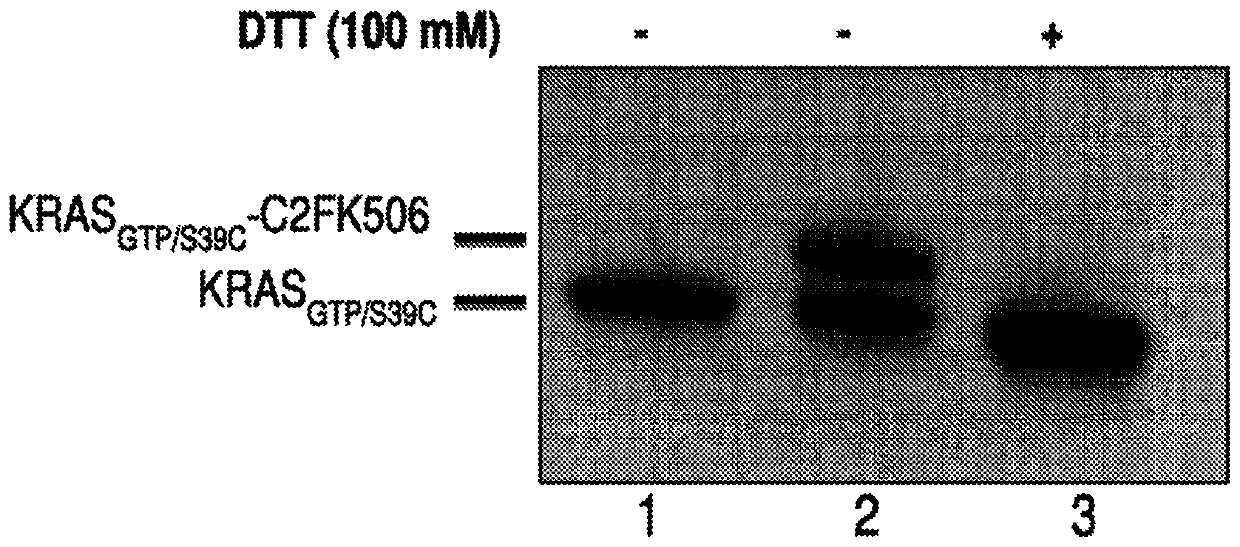
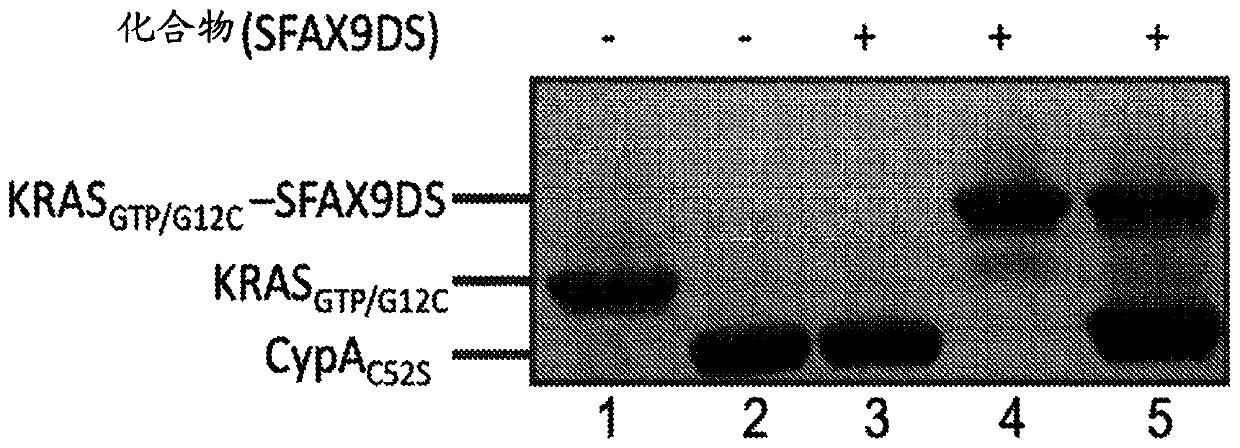
[0677] Experimental protocol: The target protein and compound were mixed together in a molar ratio of 1:2 in 12.5 mM HEPES pH 7.4, 75 mM NaCl buffer containing 2% DMSO. Reactions were incubated at 37°C for 30 min and then at room temperature overnight. Cross-linking efficiency was assessed by SDS-PAGE gel. The conjugate migrates more slowly than the non-crosslinked target protein. For thiol-reactive compounds, the Cys-specific attachment of the compound to the target protein can be further confirmed by SDS-PAGE after adding 100 mM DTT to the reaction mixture, which reduces the return of the conjugate to its individual components.
[0678] A. KRAS GTP / S39C Formation of the ...
Embodiment 3
[0690] Example 3: Formation of certain complexes
[0691] General protocol: The protocol describes two methods for the formation and isolation of complexes containing presented proteins, compounds, and mammalian target proteins.
[0692] Reagents: Compound (internal), presented protein (internal), and mammalian target protein (internal) in 100% DMSO
[0693] Equipment: Micro PROTEAN TGX gel (Bio-Rad), Superdex 75 (GE Healthcare, CV 120mL)
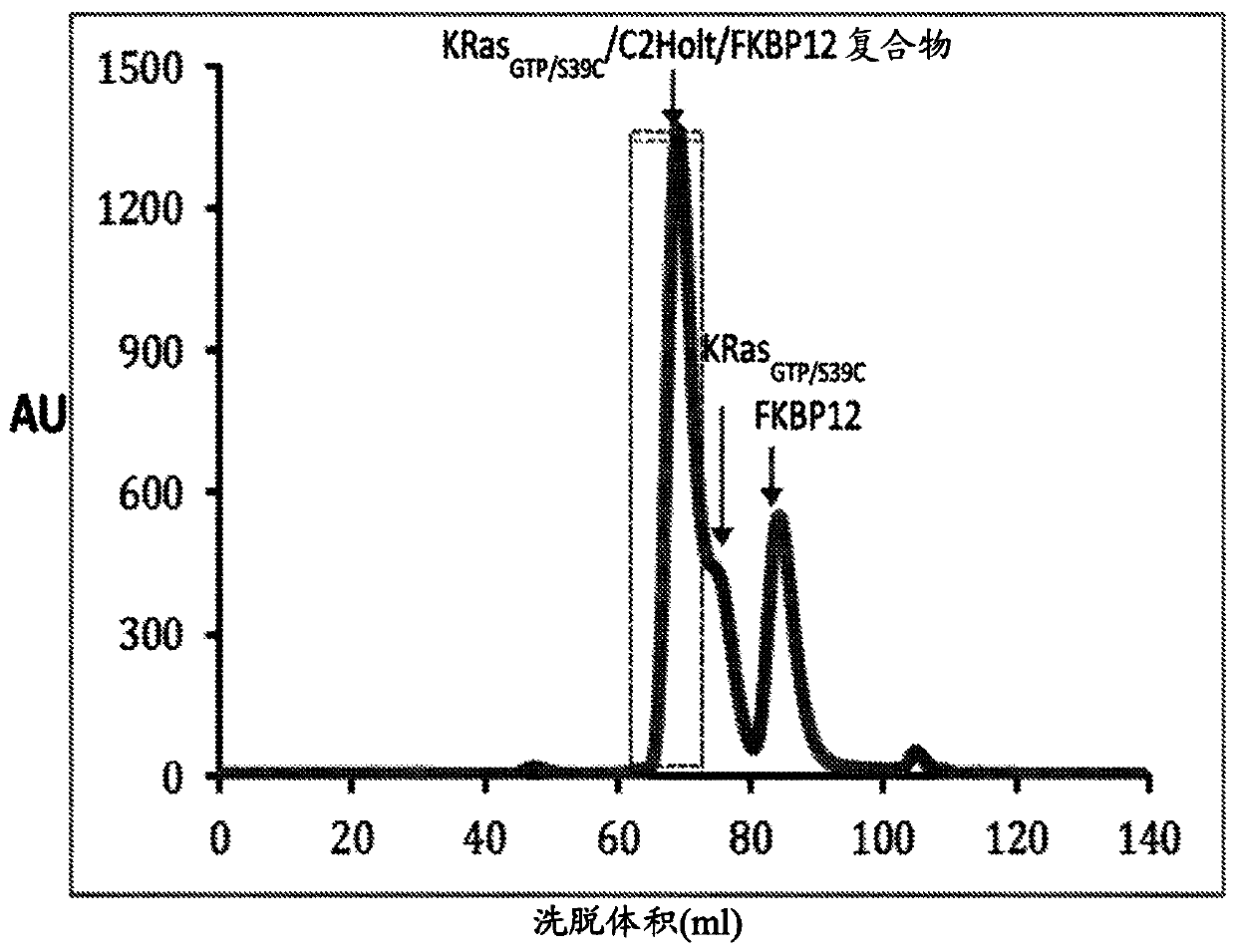
[0694] Experimental Protocol A: Preconjugated Compounds and Proteins
[0695] The conjugate and presentation protein were mixed together in a molar ratio of 1:2 in 12.5 mM HEPES pH 7.4, 75 mM NaCl buffer containing 2% DMSO. Reactions were incubated at 37°C for 30 min and then at room temperature overnight. Pure complexes were isolated by size exclusion chromatography (SEC) purification. The reaction mixture was injected directly onto a Superdex 75 column (CV 120 mL) pre-equilibrated with a buffer containing 12.5 mM HEPES pH 7.4, 75 mM N...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com