Novel base conversion editing system and application thereof
An editing system and base technology, applied in the field of gene editing, can solve the problems of narrow cytosine deaminase mutation window, inability to change, and low activity of adenine deaminase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0157] Embodiment 1, the construction of the second vector or the second nucleic acid construct
[0158] 1. Target sequences derived from human loci EMX1, PD-1, VEGFA
[0159]According to the working principle of CRISPR / Cas9 and BE3, ABE7.10, in NCBI, obtain the targets where C or A simultaneously appear in the ACBE window on human loci (such as EMX1, PD-1, VEGFA), as shown in Table 1 below shown.
[0160] Table 1. Target sequences of human-derived loci EMX1, PD-1, VEGFA
[0161] target name sequence(5`-3`) EMX1-BE3-sg1 SEQ ID NO.8 AAGGACGGCGGCACCGGCGGGGG PD-1-sg1 SEQ ID NO.9 CTGCAGCTTCTCCAACACATCGG PD-1-sg2 SEQ ID NO.10 CAGCAACCAGACGGACAAGCTGG PD-1-sg3 SEQ ID NO.11 GGACCGCAGCCAGCCCGGCCAGG PD-1-sg4 SEQ ID NO.12 CTTCCACATGAGCGTGGTCAGGG VEGFA-sg2 SEQ ID NO.13 GGCGAGCCGCGGGCAGGGGCCGG
[0162] 2. Design sgRNAoligo according to the target sequence
[0163] The construction of the target plasmid (the target p...
Embodiment 2
[0177] Embodiment 2, the construction of ABE7.10 gene editing vector or nucleic acid construct
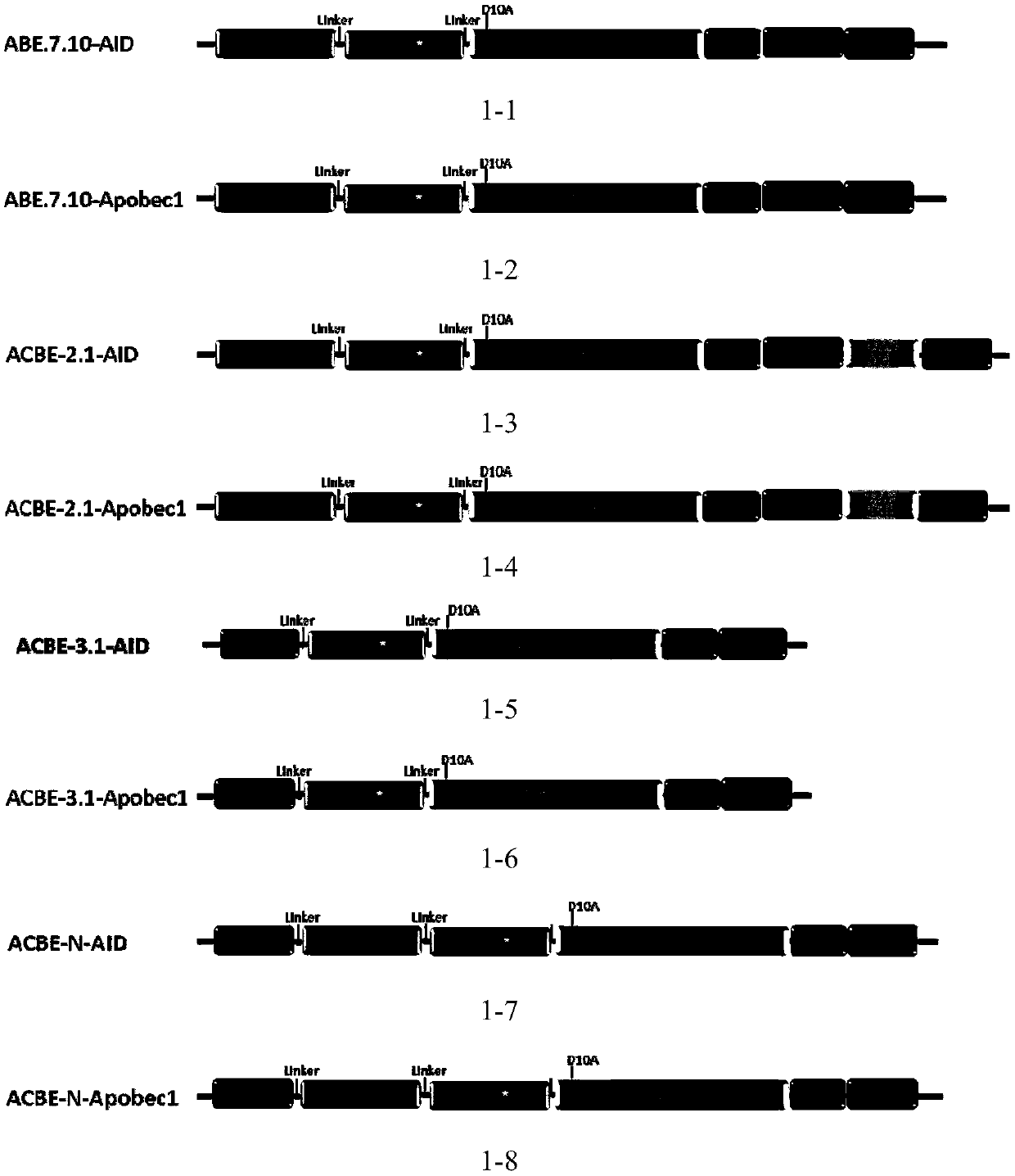
[0178] On the basis of ABE7.10 (addgene #102919), AID and Apobec1 were respectively cloned to the C-terminal of ABE7.10 by PCR, and wild TadA was cloned to the N-terminal of ABE7.10 to obtain the following Picture 1-1 and the plasmids or constructs shown in 1-2.
Embodiment 3
[0179] Example 3. Design and construction of different base conversion editing systems ACBE
[0180] 1. in Picture 1-1 and Figure 1-2 Based on the base conversion editing system, other different base conversion editing systems were designed, that is, AID and Apobec1 were respectively cloned into the C-terminal and N-terminal of ABE7. Clone a T after the C-terminus 2 A-UGI, you can get as follows figure 1 Plasmids indicated. The primers used are shown in Table 3 below:
[0181] Table 3 Primer sequences for constructing different base conversion editing systems
[0182]
[0183]
[0184] 2. Construction of U6-sgRNA-CMV-UGI-T 2 A-GFP
[0185] With the primers listed in Table 4 below, CMV, UGI, T 2 A-GFP, assembled onto U6-sgRNA(EcoRV+NotI) plasmid.
[0186] Table 4 Primer sequences for constructing different base conversion editing systems
[0187]
[0188] 3. Detection of working window and working efficiency of ACBE-N-AID and ACBE-N-Apobec1 mutant endogenou...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com