A method and indel molecular marker for traceable origin of maize mixed pollination hybrids
A technology of molecular markers and mixed pollination, which is applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of manpower consumption, process management difficulties, and influence on breeding process, and achieve stable detection results, low detection cost, The effect of detecting convenience
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Development and Validation of an InDel Molecular Marker Traceable to the Origin of Maize Mixed-pollinated Hybrids:
[0026] Download the B73 reference genome sequence (V4) from the maizeGDB (https: / / www.maizegdb.org / ) website, and download 205 maize autogenous sequences from the NCBI (https: / / www.ncbi.nlm.nih.gov / ) website Inbred genome resequencing data (PRJNA82843, SRP011907 and PRJNA260788). The genome resequencing data were subjected to Q20 (Phred-Score≥20, ie 1% error rate) quality control and sequencing length filtering (L20, length≥20bp) by SoleaQ software package. The cleaned up sequencing data were assembled de novo by SOAPdenovo2 software.
[0027] The mInDel software was used to identify the InDel variation between the assembled sequence and the B73 reference genome and to design primer sequences. Primers were selected according to the conditions of unique InDel genome location, large InDel differences, and high primer design scores, and the co-dominant mol...
Embodiment 2
[0030] A method for traceability of the source of mixed-pollinated hybrids of corn, comprising the steps of:
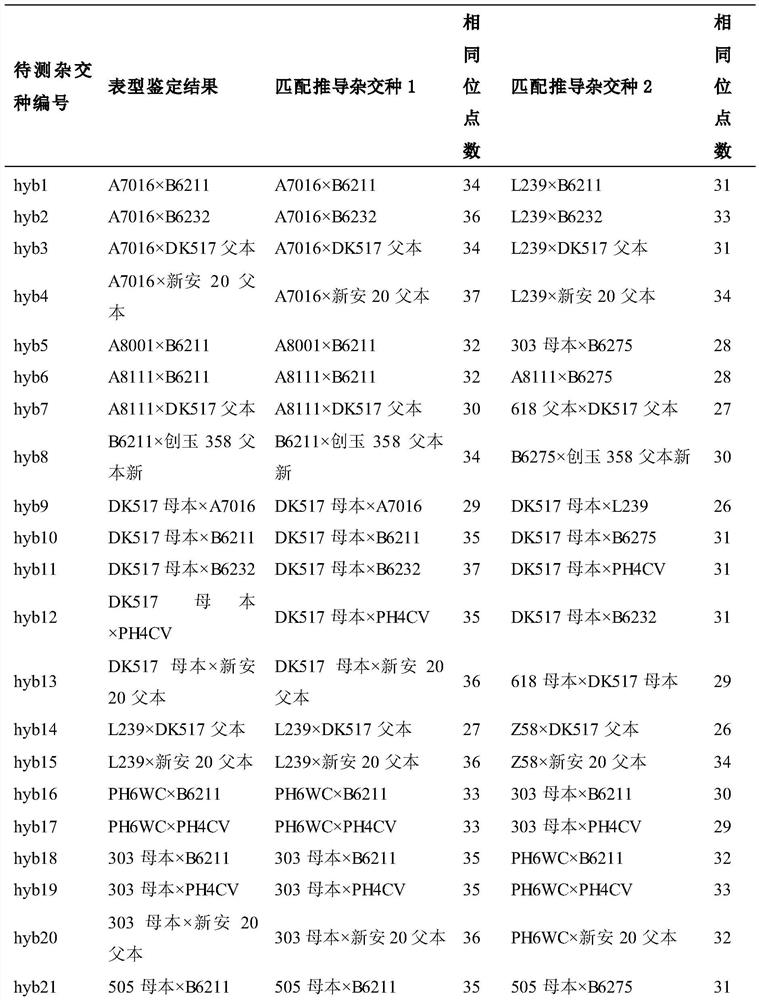
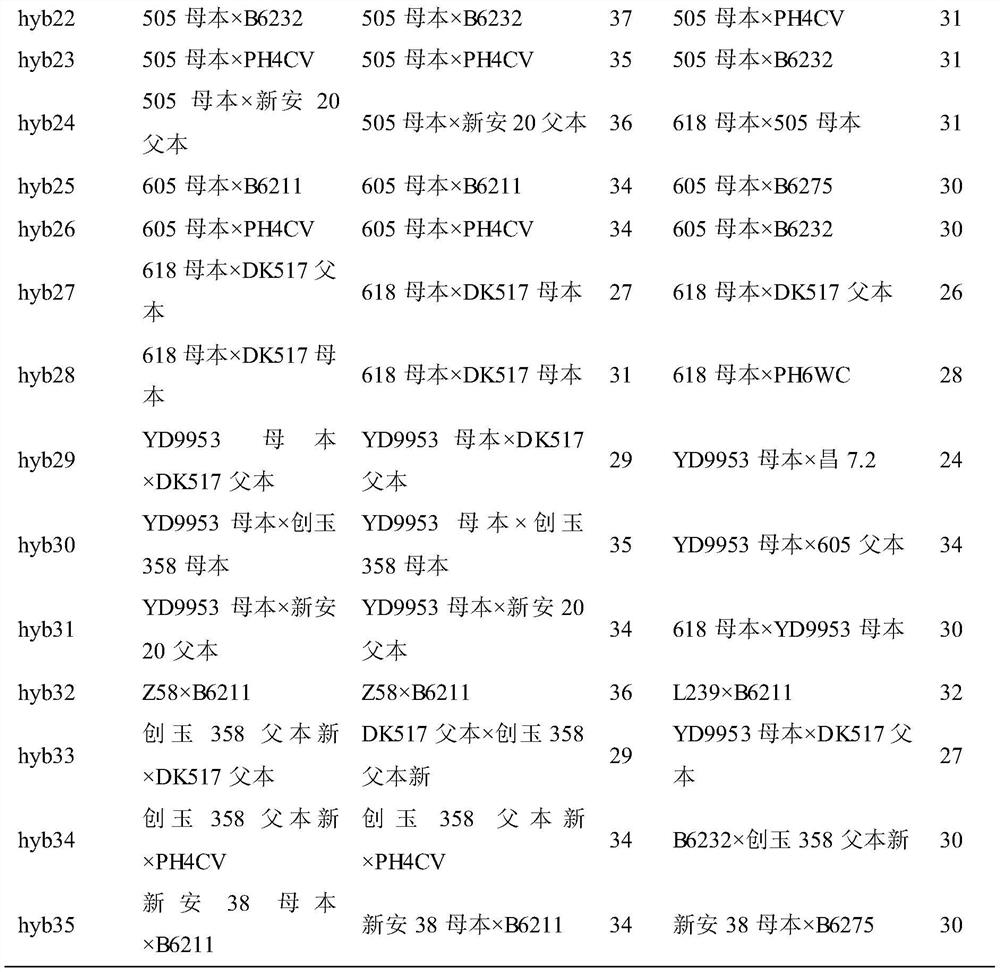
[0031] (1) Combination of hybrids to be tested:
[0032] Twenty-five parental inbred lines were planted in the experimental field, and the tassel was removed from the female parent row before tasseling. The pollination method is natural open pollination in the field. The ear after pollination is listed for harvest, and after threshing, it is planted as a single seed, and the parent is planted at the same time. Seeds from the same ear were planted together for later identification of hybrid male parents by phenotypic comparison.
[0033] (2) Obtaining the genotypes of the parents and the hybrid to be tested:
[0034] Using the 37 high-quality InDel molecular markers verified in Example 1, 25 parental inbred lines and 49 randomly selected hybrids to be tested were genotyped by agarose gel electrophoresis. The short band of each molecular marker was recorded as A, th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com