Molecular marker closely linked with major QTL (quantitative trait loci) of lateral root number in wheat seedling stage as well as detection primer and application of molecular marker
A molecular marker, number-based technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problem that the number of lateral root molecular markers cannot be effectively used in molecular-assisted breeding, and the linkage markers are far from the target gene. and other problems, to achieve the effect of promoting transfer application, fast and efficient detection, and stable amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1. Molecular markers closely linked to the major QTL for the number of lateral roots in wheat seedling stage
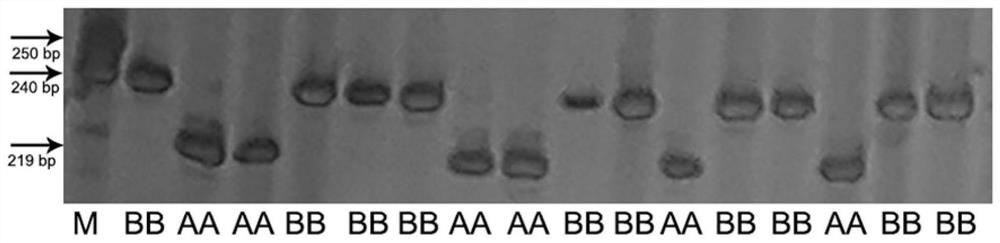
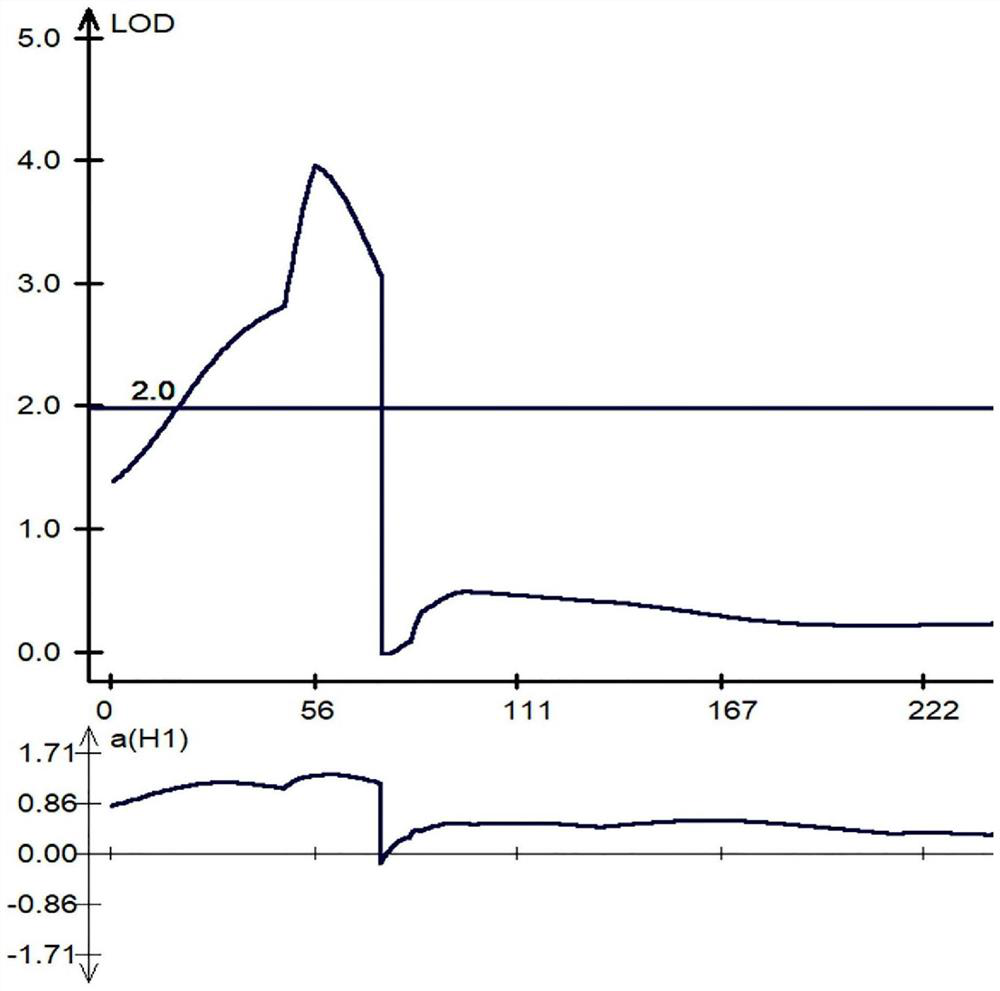
[0037]Using common wheat TAA10 and synthetic hexaploid wheat XX329 as parents, 182 RIL families were derived as test materials. Using QTL mapping analysis, the main QTL for the number of lateral roots (QLrn5D) was located on chromosome 5D. Using TAA10 and XX329 Based on the resequencing data, an InDel molecular marker QAU5D-20 with polymorphism among the parents was designed and developed near the LOD prediction peak of the QLrn5D segment. The sequence of the upstream primer of the molecular marker QAU5D-20 is: TTGATAAAAAGCCCCCTTCC (SEQ ID NO: 2), and the sequence of the downstream primer is: GCTAATTAGACCCGGCAACA (SEQ ID NO: 3).
[0038] Using the genomic DNA of wheat variety TAA10 as a template, the upstream primer shown in SEQ ID NO:2 and the downstream primer shown in SEQ ID NO:3 were used for PCR amplification.
[0039] The PCR amplification reacti...
Embodiment 2
[0049] Example 2, Acquisition of Molecular Marker QAU5D-20
[0050] The method for obtaining the molecular marker QAU5D-20 specifically includes the following steps:
[0051] Step 1: Form the F8 generation RIL population of 182 families
[0052] Hybrid F was obtained by crossing TAA10 and XX329 as parents 1 , hybrid F 1 Selfing produces F2,
[0053] f 2 The F8 generation RIL population containing 182 families was formed by selfing generation by generation;
[0054] Step 2: extracting leaf DNA of wheat RIL population with CTAB method, specific methods and procedures are as follows:
[0055] (1) Take 0.2 g of young leaves of wheat plants, put them in a 1.5 ml Eppendorf tube, add liquid nitrogen, and grind them into fine powder.
[0056] (2) Add 600 μl of 1×CTAB extraction buffer preheated at 65°C to the centrifuge tube, and shake gently to mix.
[0057] (3) Incubate in a 65°C water bath for 60 minutes and shake the centrifuge tube carefully every 10 minutes.
[0058] (4)...
Embodiment 3
[0092] Embodiment 3, the application of molecular marker QAU5D-20
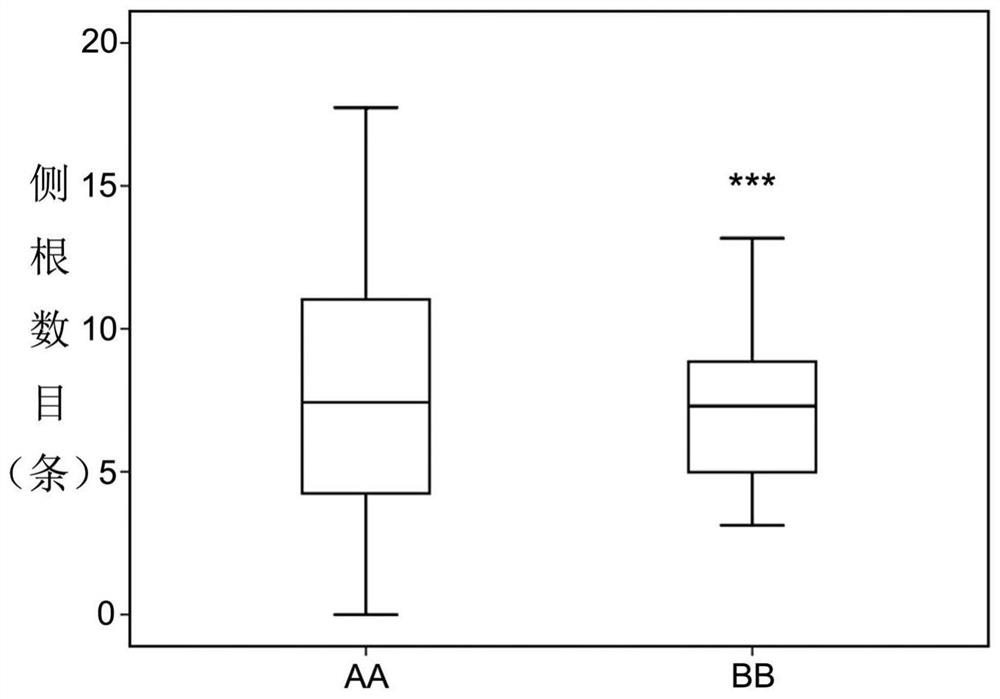
[0093] Based on previous tests and analysis, it can be seen that: (1) the primer pair consisting of the upstream primer shown in SEQ ID NO: 2 and the downstream primer shown in SEQ ID NO: 3 can be used in identification or auxiliary identification of the number of lateral roots in wheat seedling stage Among traits, it can also be used to obtain molecular markers related to the number of lateral roots in wheat seedling stage; (2) Molecular marker QAU5D-20 can be used in identifying or assisting in identifying the number of lateral roots in wheat seedling stage, and can also be used in breeding Lateral root number genotypes in wheat.
[0094] In summary, the primer pair and molecular marker QAU5D-20 provided by the present invention can greatly accelerate the molecular marker-assisted breeding process of wheat lateral root number, and have important application value for genetic improvement of wheat root system ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com