Primer group and kit for amplifying PKD1 gene
A technology of primer sets and kits, applied in the field of genetic detection of genetic diseases, can solve the problems of cumbersome methods, low detection efficiency, high workload and cost, and achieve accurate detection results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] Embodiment 1 Long fragment PCR amplification
[0085] 1. Genomic DNA Extraction
[0086] Collect human peripheral blood, use Tiangen blood extraction kit to extract whole genome gDNA, measure DNA concentration with Nanodrop and Qubit3.0 instrument, record the corresponding concentration and the ratio of 260 / 280 and 260 / 230.
[0087] 2. Long fragment PCR amplification
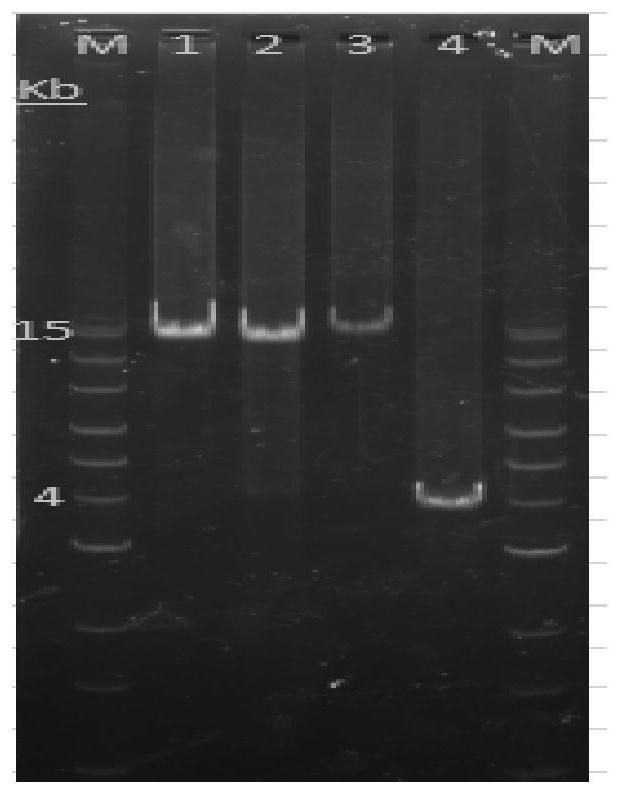
[0088] Using the whole genome gDNA extracted above as a template, all PKD1 genes were amplified according to the primer pairs in Table 1 below, the reaction systems in Table 2 and Table 3, and the amplification programs in Table 4 and Table 5. In the primer names in Table 1, F represents the upstream primer (forward primer), and R represents the downstream primer (reverse primer). The kit used for long fragment PCR amplification is KFX-201 (200U×1), which consists of KOD FX Noe (1.0U / μL), 2×PCR Buffer for KOD FX Neo, 2mM dNTPs, manufacturer: TOYOBO. Reaction system 1 in Table 2 is suitable for amplific...
Embodiment 2
[0102] Embodiment 2 library construction
[0103] 1. Purification of long fragment PCR products
[0104] PCR products were purified using Yisheng or Novizan DNA purification magnetic beads 1.0×sample volume, washed twice with 80% ethanol, and 30 μL Nuclease-free ddH 2 O was eluted, and its concentration was determined by the Qubit dye method.
[0105] 2. The purified product is used for library construction with Yisheng library construction kit
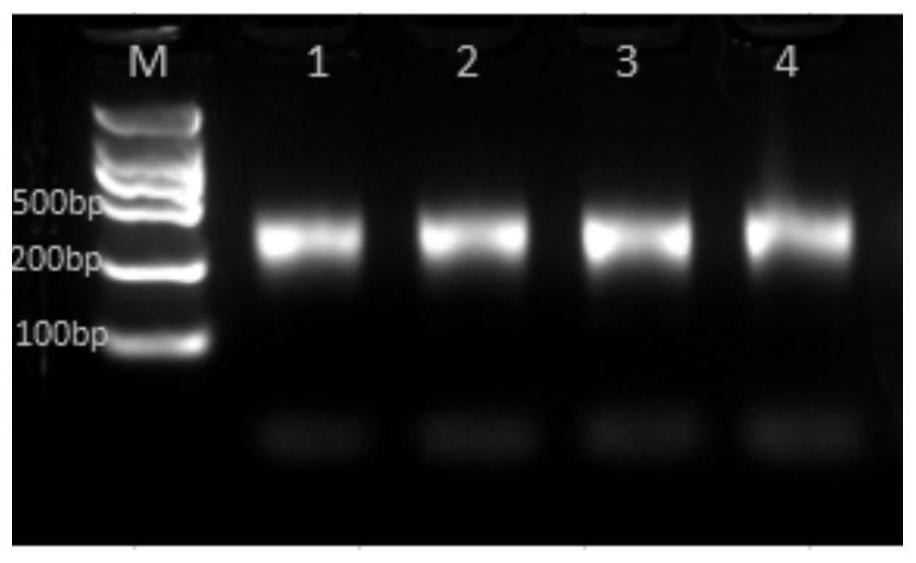
[0106] (1) DNA fragmentation: 120 ng of purified PKD1 four fragments were taken separately for use.
[0107] Use Yisheng rapid fragmentation / end repair / A tail addition module reagents to prepare a DNA fragmentation reaction system according to Table 6, and perform the PCR reaction procedure in Table 7 after the preparation is completed.
[0108] Table 6 DNA fragmentation reaction system
[0109] Reagent name Volume (μL) Purified product DNA (120ng) x Smearase Mix 10 Nuclease-free ddH 2 o
(50-x) ...
Embodiment 3
[0129] Embodiment 3 data analysis
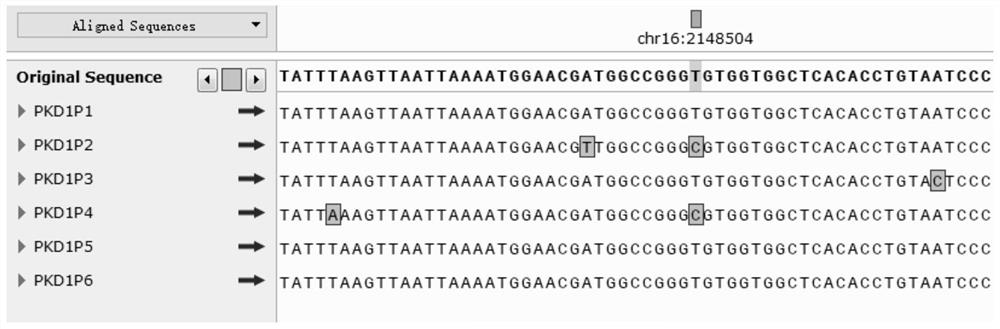
[0130] First, the original fastq file of the PKD1 gene (fastq file: the data returned by sequencing, mainly sequence information and corresponding quality value) is filtered to remove the data with poor quality, and then compared with the pseudogene locus library to detect each The pseudogene-specific sites of the amplicon library in the four amplicon regions respectively. If a pseudogene-specific site is detected, the ratio of the pseudogene bases in the site to the coverage depth of all bases is calculated, which is called The pseudogene frequency at that locus is calculated as the average of the pseudogene frequencies for all pseudogene-specific loci within each amplicon.
[0131] The results of pseudogene-specific site analysis are shown in Table 12, which is the detected pseudogene-specific site information in different amplicon library data.
[0132] Table 12 Pseudogene-specific site analysis results
[0133]
[0134] As shown in th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com