Nucleic acid linker for high-throughput sequencing and library construction method
A technology for sequencing library and construction method, applied in the field of high-throughput sequencing, can solve the problems of long time consumption, adapter dimer pollution, high cost, and achieve the effect of reducing adapter self-ligation, reducing adapter self-ligation, and reducing self-ligation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
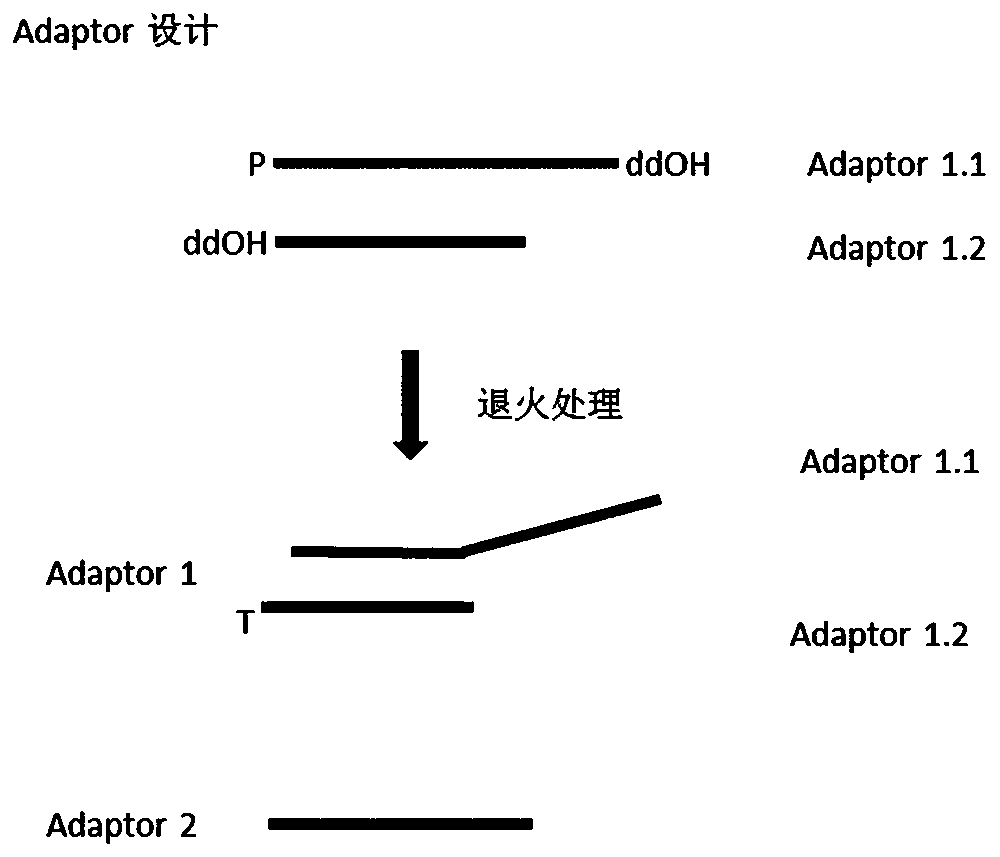
[0053] Example 1 Adapter for high-throughput sequencing (1)
[0054] This embodiment provides a linker for high-throughput sequencing, which consists of linker 1 and linker 2. Linker 1 is a double-stranded oligonucleotide of unequal length, and its long-chain sequence is shown in SEQ ID NO.1. The short-chain The sequence is shown in SEQ ID NO.2, the 5' end of the long chain is modified by phosphorylation, the 3' end of the long chain and the 3' end of the short chain are modified by dideoxy; the linker 2 is a single-stranded oligonucleotide, and the sequence is as follows As shown in SEQ ID NO.3, linker 2 contains a region complementary to the 5' end of the long chain of linker 1.
[0055] The above-mentioned high-throughput sequencing adapters can be used for the construction of sequencing libraries on the BGI sequencing platform.
Embodiment 2
[0056] Example 2 Adapter for high-throughput sequencing (2)
[0057] This embodiment provides a linker for high-throughput sequencing, which consists of linker 1 and linker 2. Linker 1 is a double-stranded oligonucleotide of unequal length, and its long-chain sequence is shown in SEQ ID NO.4. The short-chain The sequence is shown in SEQ ID NO.5, the 5' end of the long chain is modified by phosphorylation, the 3' end of the long chain and the 3' end of the short chain are modified by dideoxy; the linker 2 is a single-stranded oligonucleotide, and the sequence is as follows As shown in SEQ ID NO.6, linker 2 contains a region complementary to the 5' end of the long chain of linker 1.
[0058] The above-mentioned high-throughput sequencing adapters can be used for sequencing library construction on the Illumina sequencing platform.
Embodiment 3
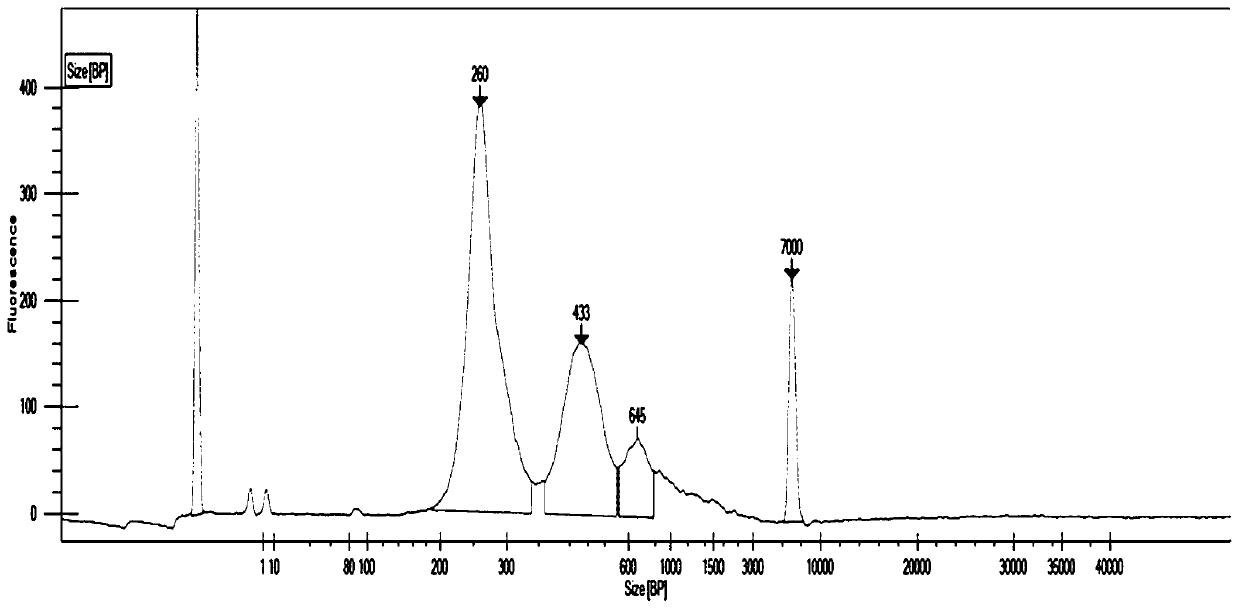
[0059] The construction of embodiment 3 sequencing library
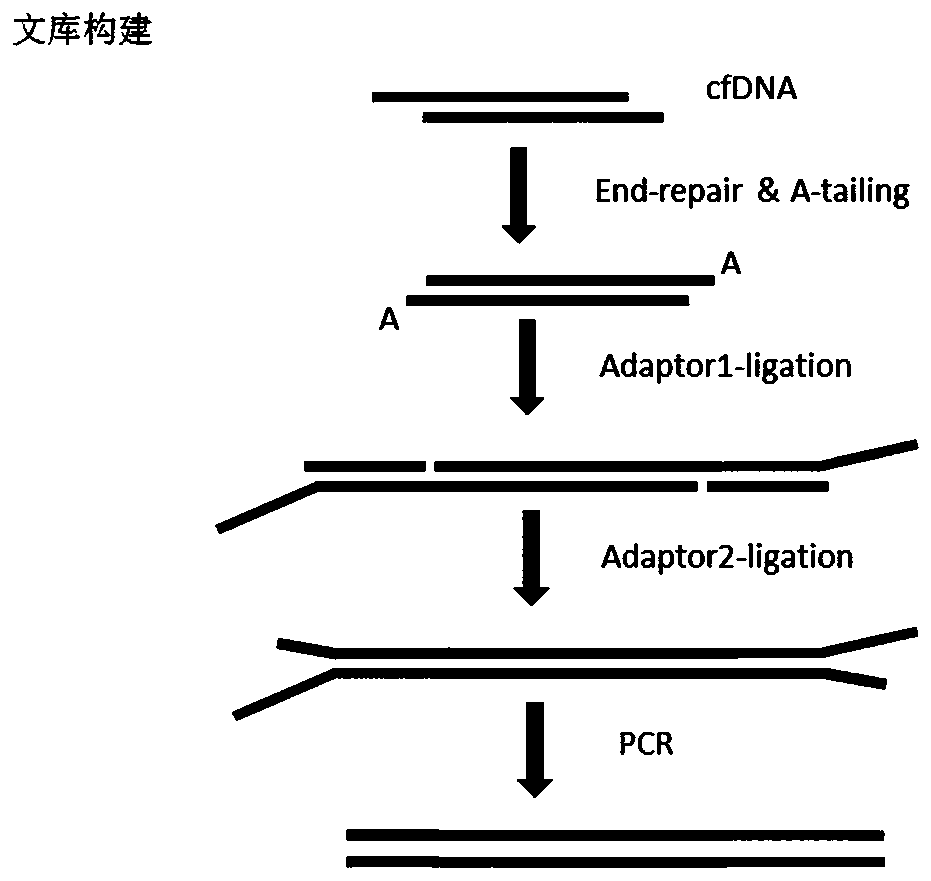
[0060] In this example, cfDNA is used as the sequencing sample (parallel experiment with 3 samples), and the adapter of Example 1 is used to construct the sequencing library. The flow chart is as follows figure 2 As shown, the specific method is as follows:
[0061] 1. End repair and addition of A: The input amount of cfDNA is 2ng, and the reaction system is shown in Table 1; the reaction program: 65°C, 20min.
[0062] Table 1 End repair and A reaction system
[0063] Reagent Volume (μl) cfDNA 15 Taq enzyme 1 10x reaction buffer 2 dNTPS 1 h 2 o
1
[0064] 2. Joint connection: connect joint 1 and joint 2 in two steps, as follows:
[0065] The end-repaired and A-added product obtained in step 1 was connected to Adapter 1, and the reaction system was shown in Table 2; reaction program: 20°C, 15min.
[0066] Table 2 Connection reaction system of adapter 1
[0067] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com