Methods to determine sensitivity profile of bacterial strain to therapeutic composition
A therapeutic composition and sensitivity technology, applied in the field of determining the sensitivity spectrum of bacterial strains to therapeutic compositions, can solve problems such as treatment delay and complications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0142] Example 1: Phage Isolation / Characterization from Environmental Sources.
[0143] Powdered TSB medium (Becton, Dickinson and Company) can be mixed with raw sewage to a final concentration of 3% w / v. Different bacterial strains can be grown to exponential phase and 1 mL of each strain is added to a 100 mL aliquot of the TSB-sewage mixture and incubated overnight at 37°C and 250 rpm. The next day, 1 mL of the infected TSB-sewage mixture was harvested and centrifuged at 8,000 xg for 5 minutes to pellet cells and debris. Transfer the supernatant to a sterile 0.22 µm Centrifuge tube filters (Coming, NY) and centrifuge at 6,000 xg to remove any residual bacteria. A 10 μL aliquot of the filtrate was mixed with 100 μL of an exponentially growing culture of the bacterial strain, incubated at 37°C for 20 minutes, mixed with 2.5 mL of molten top agar (0.6% agar) tempered to 50°C, and poured over TSB agar plate (1.5% TSB agar). Plates were incubated overnight at 37°C and subseq...
Embodiment 2
[0147] Example 2: Assays for generating phage-host susceptibility profiles.
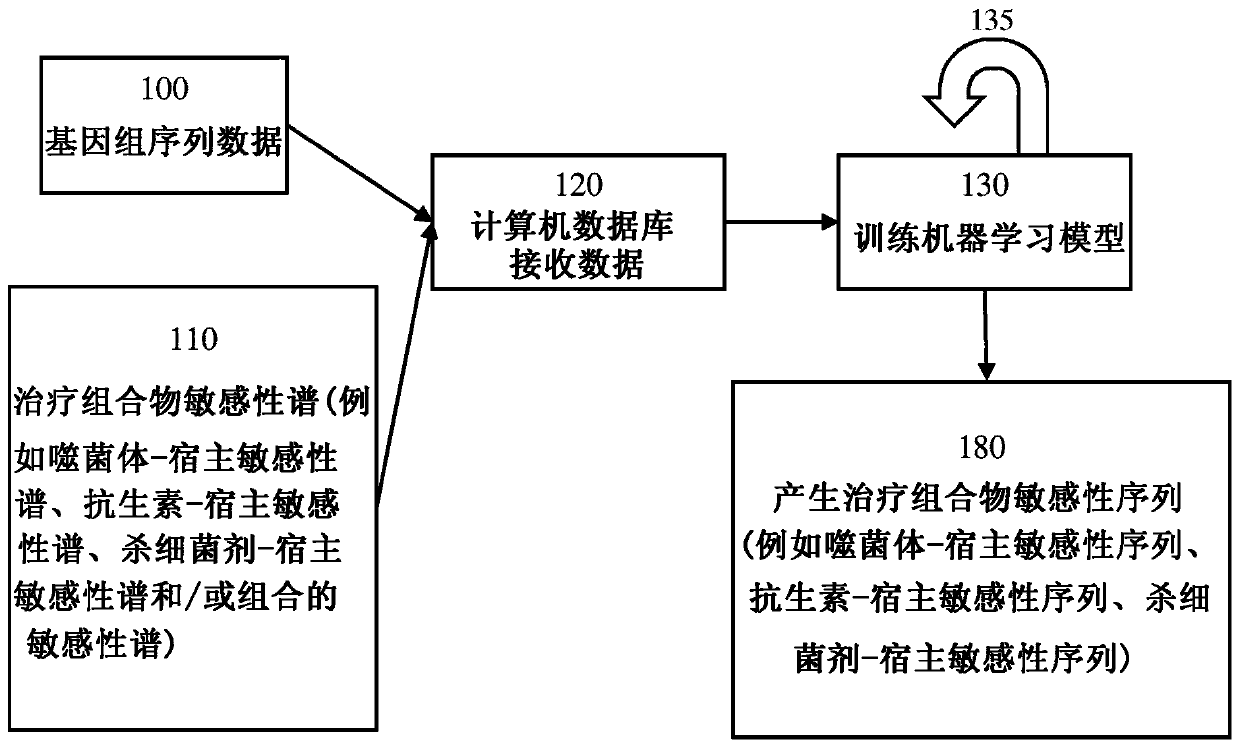
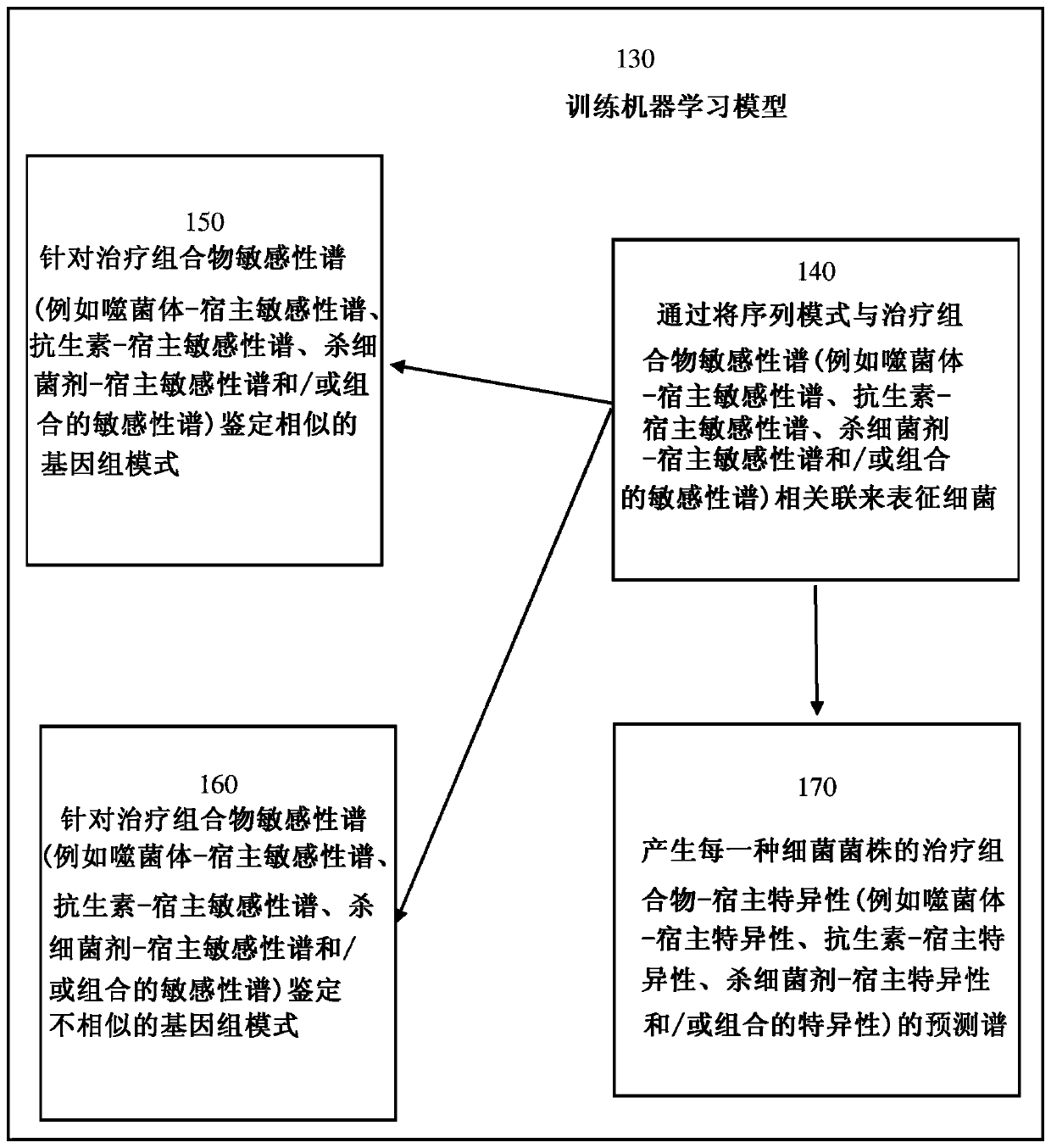
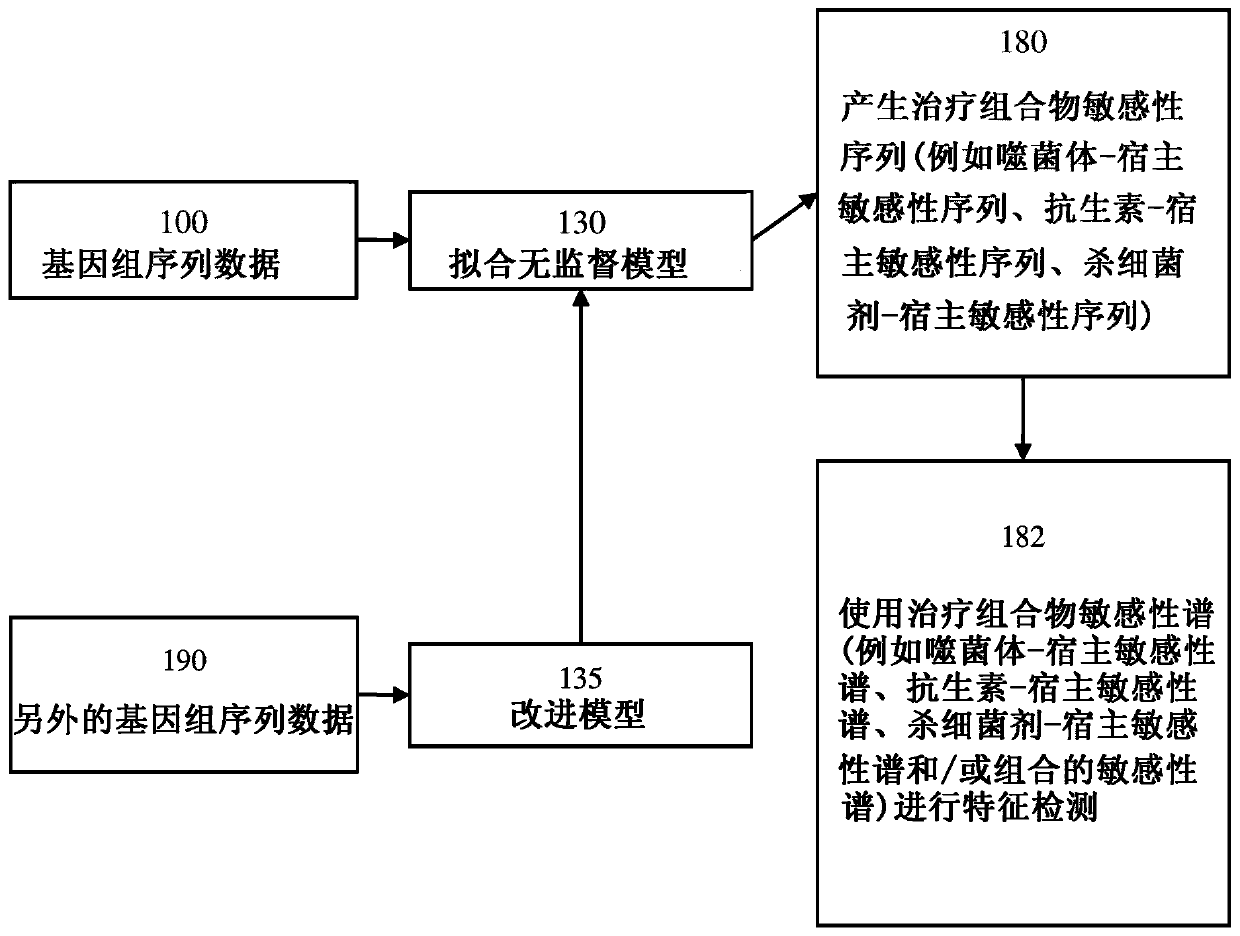
[0148] In order to perform the disclosed methods, it is necessary to compare the genomes of multiple different bacterial strains with similar or identical phage-host susceptibility profiles. If the phage-host susceptibility profile of the bacteria is known, then the following assays need not be performed. However, if the phage-host susceptibility profile of the bacterium is unknown, such a profile can be determined or experimentally derived using any of the following assays.
[0149] One method of determining the susceptibility / resistance profile of bacteria relies on automated indirect liquid lysis assays. Briefly, overnight cultures of bacterial strains were inoculated with 1% v / v tetrazole Dye mix wells of TSB 96-well plate. Phage was then added to each well, and the plate was placed in the OmniLog TM System (Biolog, InC, Hayward, CA) was incubated overnight at 37°C. See Henry, Bacteriophage 2...
Embodiment 3
[0156] Example 3: Genome Sequencing, Assembly and Annotation
[0157] Phage and / or bacterial genomes can be sequenced using standard sequencing techniques and assembled using contig analysis well known in the art. For example, 5 μg of DNA isolated from phage or bacteria can be extracted and shipped to a contract sequencing facility. 40x to 65x sequencing coverage was obtained using pyrosequencing technology on a 454FLX instrument. Files generated by the 454FLX instrument were assembled with GS assembler (454, Branford, Conn.) to generate consensus sequences. Quality improvement of the genomic sequence may involve sequencing 15-25 PCR products across the genome to ensure correct assembly, double-strand formation, and resolution of any remaining base conflicts occurring within identical polynucleotide tracts. The open reading frame (ORF) of the encoded protein can be predicted using standard programs known in the art (eg, BLASTP), followed by manual evaluation and correction i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com