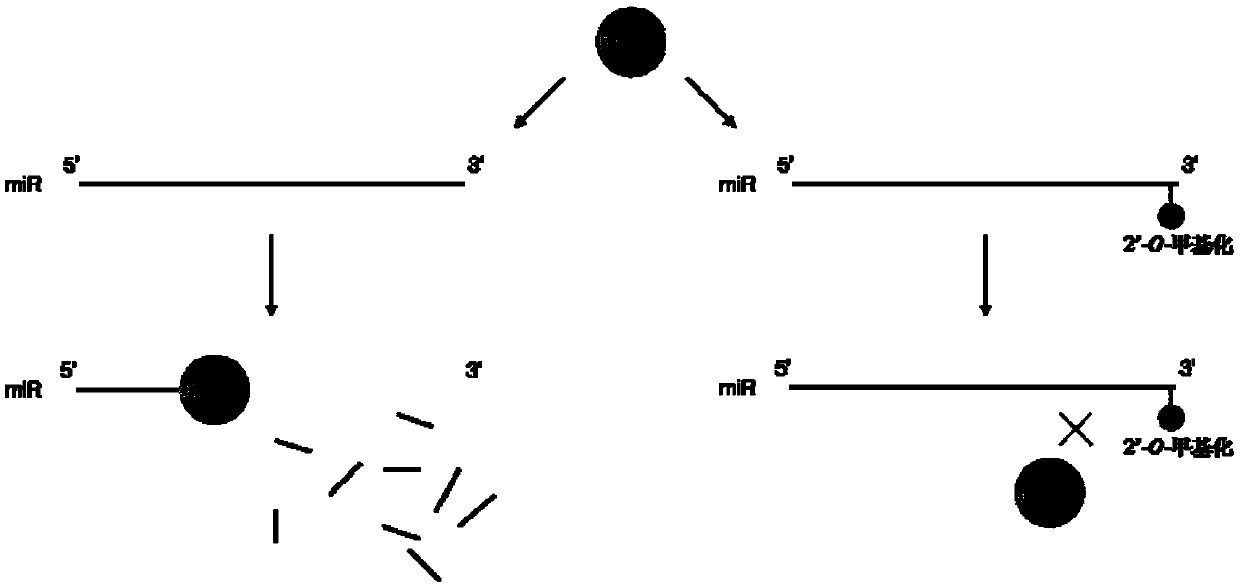
Method for identifying 2'-O-methylation modification in RNA molecules and application thereof
A methylation and molecular technology, applied in biochemical equipment and methods, analytical materials, biological material analysis, etc., can solve the problems of large sample requirements and low sensitivity, and achieve high sensitivity, simple operation, and mild reaction conditions Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Embodiment 1 recombinant protein expression and purification
[0053] Entrust Invitrogen to synthesize the RNase R gene sequence and remove the first 81 amino acid truncated gene sequences (SEQ ID NO: 1 and 2), and introduce NcoI endonuclease site at the 5' end of the gene fragment, and introduce HindIII at the 3' end endonuclease site. The synthesized gene fragment and the pET28a(+) vector were digested with NcoI and HindIII, respectively, and the gene fragment and the vector fragment were ligated using T4DNA ligase, and routinely transformed into DH5α competent cells (Tiangen Biochemical Technology (Beijing) Co., Ltd.). Positive clones were screened according to kanamycin resistance, and plasmids were extracted. The recombinant plasmid was identified by NcoⅠ and HindⅢ double enzyme digestion and agarose gel electrophoresis. Invitrogen Company was entrusted to sequence the recombinant plasmid. BioEdit software was used to analyze the sequencing results. The result was...
Embodiment 2
[0063] The preparation of embodiment 2 small molecule RNA substrates
[0064] Commissioned Invitrogen to synthesize single-stranded small RNA (miR-21) (SEQ ID NO: 5) with the same sequence: one without any additional modification (miR-21), one with 2'-O at the 3' end - Methylation modification (miR-21-ch3). They were diluted to 100 nmol and stored at -20°C in the dark.
[0065] SEQ ID NO: 5 artificially synthesized single-stranded small RNA UAGCUUAUCAGACUGAUGUUGA
Embodiment 3
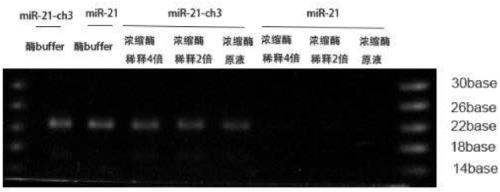
[0066] Example 3 Rnase R enzyme degradation reaction and electrophoresis detection
[0067] Mix 10 μg, 5 μg, or 2.5 μg of the recombinant fusion protein obtained in Example 1 with 7 μL of the diluted RNA substrate obtained in Example 2, add an appropriate amount of buffer solution and water to a final volume of 10 μL (20 mM Tris pH 8.5, 100 mM KCl ,0.01mM ZnCl 2 ), react at 37°C for 1 hour, and then treat at 85°C for 5 minutes to inactivate the enzyme. Afterwards, the product was separated on a 10% TBE-urea gel.
[0068] see results figure 2 . In the control group using enzyme buffer, the RNA substrates were clearly displayed on the gel regardless of whether the 3' end was modified or not. After reacting with three different concentrations of RNase R enzyme, the 2'-O-methylated RNA substrate at the 3' end was also clearly displayed on the gel. After reacting with three different concentrations of RNase R enzyme, the RNA substrate without modification at the 3' end has be...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com