PEDV genome analysis method based on next-generation sequencing
A virus genome and analysis method technology, applied in the field of PEDV virus genome analysis based on next-generation sequencing, can solve problems such as inability to accurately identify the gene structure and affect PEDV virus genome splicing, and achieve the effect of avoiding omissions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
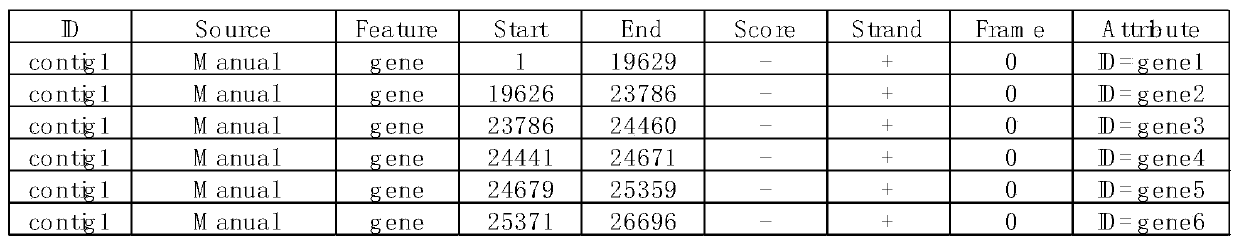
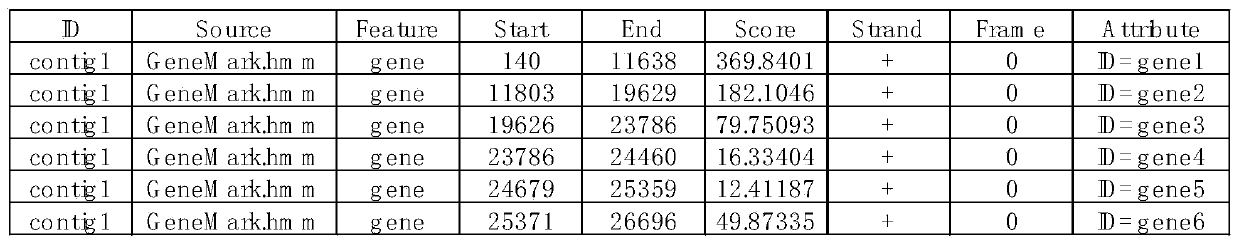
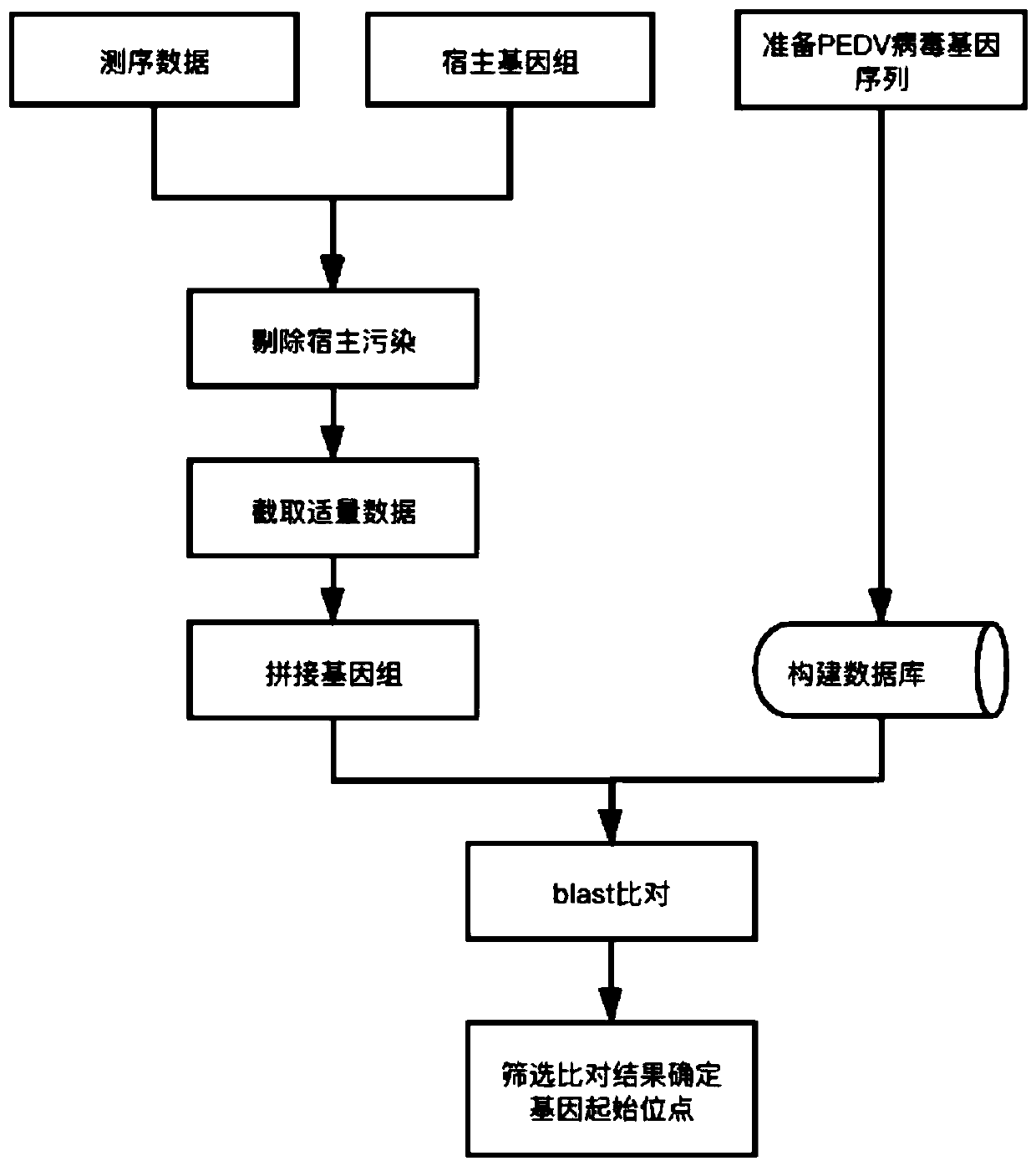
[0024] 1. The PEDV virus sample pedv1 was passed through the illumina Miseq sequencer to generate 2.4G of raw data, and 2.3G of high-quality sequences in fastq format were obtained after removing sequencing adapters and quality control.
[0025] 2. Since the host of the PEDV virus is a pig, download the reference genome and transcript sequence of the pig from the Ensembl website.
[0026] 3. Run bowtie2-build to build an index with the reference sequence downloaded in step (2) as input.
[0027] 4. Run the Bowtie2 software (default parameters) to compare the sequencing data with the host reference sequence in step (1).
[0028] 5. According to the comparison results, the data compared to the pig reference sequence were eliminated. Obtain sequence sequences free of host contamination.
[0029] 6. Since too much data volume is not conducive to splicing, the sequencing sequence without host contamination obtained in step (5) intercepts 4.2m of data volume about 150 times the si...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com