Cell strain capable of stably expressing AMPA receptor GluR1/GluR2, and construction method of cell strain
A construction method and stable expression technology, applied in the field of genetic engineering, can solve problems such as loss of exogenous genes in cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
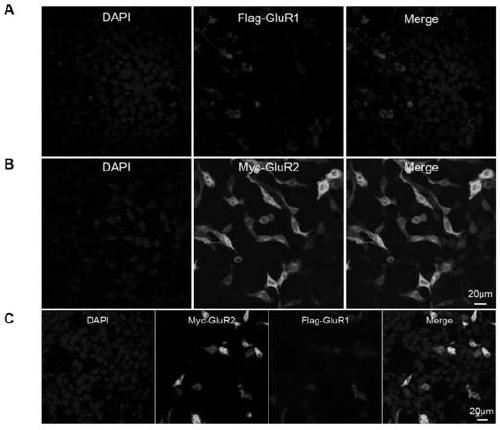
[0040] Example 1 Transient expression verification
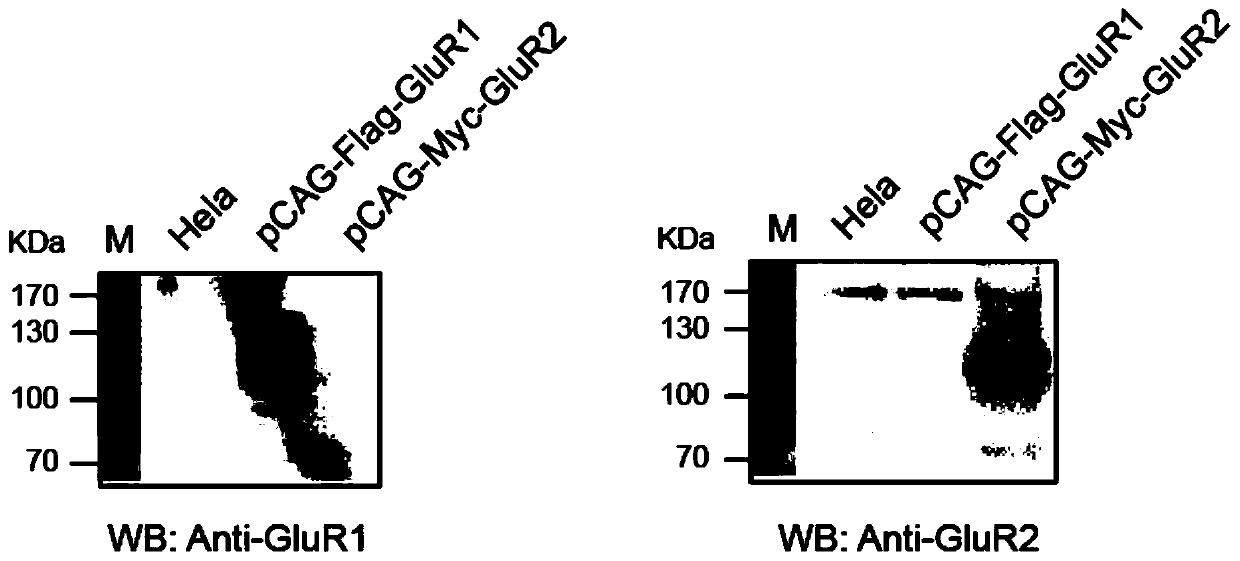
[0041] By constructing pCAG-3×Flag-GluR1 and pCAG-Myc-GluR2 vectors, PEI transfection reagents were used to transfect Hela cells, so that the target gene was expressed in a short but high level in Hela cell lines, and then the cells were detected by immunofluorescence technology. Localize the target protein expressed in the cell to detect whether the target protein is expressed correctly.
[0042]Using the pCAG-pHluorin-GluR1-Yong plasmid as the backbone, EcoRI and KpnI endonuclease cleavage treatment removes the pHluorin protein to form a linear vector with cohesive ends. The pCAG-3×Flag-GluR1 and pCAG-Myc-GluR2 vectors were constructed by PCR amplification; the constructed plasmids were transfected into Hela cells with PEI (Polysciences, USA) transfection reagent, and cultured at 37°C and 5% CO2 for 48 After 1 hour, the total cell protein was collected, and the expression of GluR1 and GluR2 was detected by Western Blot. ...
Embodiment 2
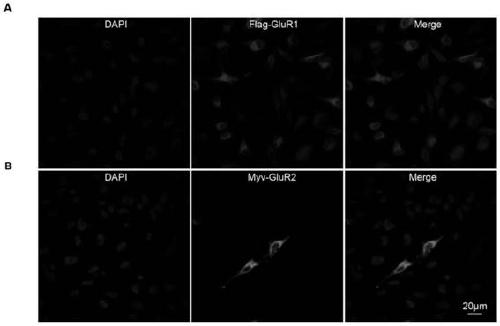
[0044] Embodiment 2 Construction of stable expression vector
[0045] The pQCXIH plasmid was used as the backbone vector, and the pCAG-3×Flag-GluR1 and CAG-Myc-GluR2 constructed above were used as templates to construct the pQCXIH-3×Flag-GluR1 and pQCXIH-Myc-GluR2 lentiviral retroviral vectors.
[0046] According to the CDS sequence of GluR1 and GluR2 and the multiple cloning site of the vector pQCXIH, primers retaining restriction sites (Not I and BamH I) are designed, and the signal peptide (GluR1 Signal peptide sequence: ATGCCGTACATCTTTGCCTTTTTCTGCACCGGTTTTTAGGTGCGGTTGTGGGTGCCAAT; GluR1 signal peptide sequence: ATGCAAAAGATTATGCATATTCTGTCCTCCTTTCTCCTGTTTTATGGGGACTGATTTTTGGTGTCTCTGCGCGC). After retroviral packaging of the constructed pQCXIH-3×Flag-GluR1 and pQCXIH-Myc-GluR2 recombinant vectors, the obtained pseudovirus was used to infect Hela cells, and 250ng / μl hygromycin B was added to screen to obtain stable expression AMPA receptor GluR1 / GluR2 cell line.
[0047] The sp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com