Primer design method and system based on k-mer algorithm
A design method and primer design technology, applied in the field of primer design method and system based on k-mer algorithm, can solve the problem of long design time, achieve high coverage, and improve the effect of primer design time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] This embodiment is a primer design method based on the k-mer algorithm, which design method includes:
[0058] (a) construct functional gene nucleic acid sequence database, and based on k-mer algorithm, be k with primer length, and k is 17-20bp, the nucleic acid sequence in the database is respectively cut into k-mers;
[0059] (b) Select 120 k-mers according to the occurrence frequency of k-mer from high to low, and use them as primers to select k-mers;
[0060] (c) Merge the k-mers with overlap > 10 among the alternative primer k-mers, and then select 40 k-mers from the combined primer alternative k-mers according to the frequency from high to low as preliminary primers, The merging method is: retain the k-mer with the highest frequency among the k-mers with overlap > 10, if there are multiple k-mers with the highest frequency, keep the longest k-mer;
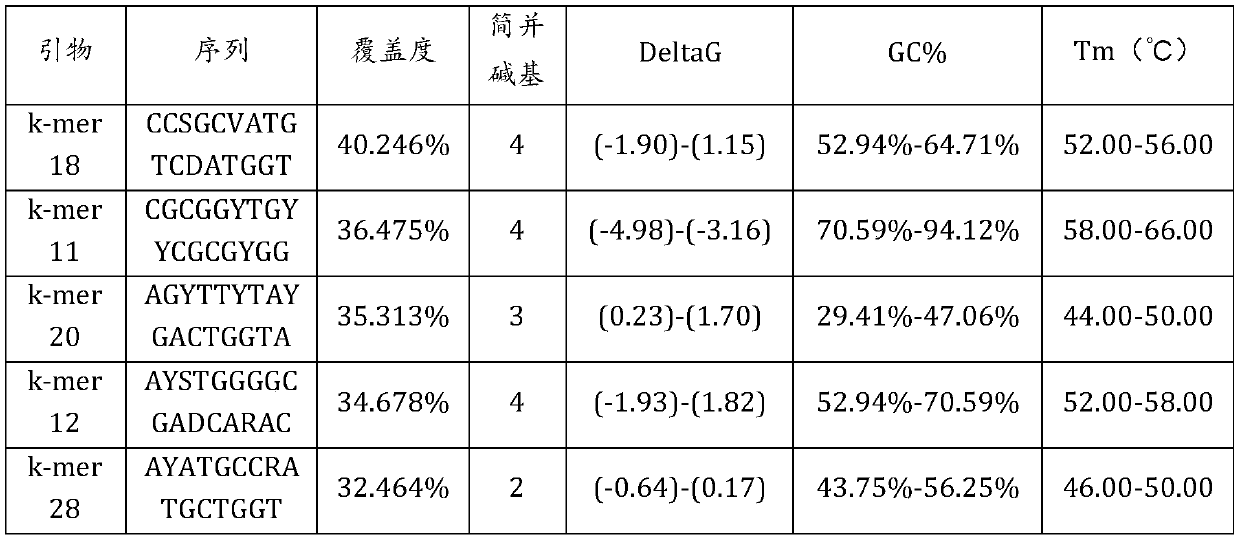
[0061] (d) Search for k-mers with a difference of one base from each preliminary primer among the k-mers with cover...
Embodiment 2
[0068] This embodiment is a primer design method based on the k-mer algorithm, which design method includes:
[0069] (a) Construct a functional gene nucleic acid sequence database, supplement the species information in the functional gene nucleic acid sequence database by gene numbering, and based on the k-mer algorithm, use the primer length as k, and k is 20bp, cut the nucleic acid sequences in the database into k-mers;
[0070] (b) Select 100 k-mers according to the occurrence frequency of k-mers from high to low, and use them as primers to select k-mers;
[0071] (c) Merge k-mers with overlap > 10 in the alternative primer k-mers, and then select 30 k-mers from high to low frequency among the combined primer alternative k-mers as preliminary primers, The merging method is: retain the k-mer with the highest frequency among the k-mers with overlap > 10, if there are multiple k-mers with the highest frequency, keep the longest k-mer;
[0072] (d) Search for k-mers with a d...
experiment example
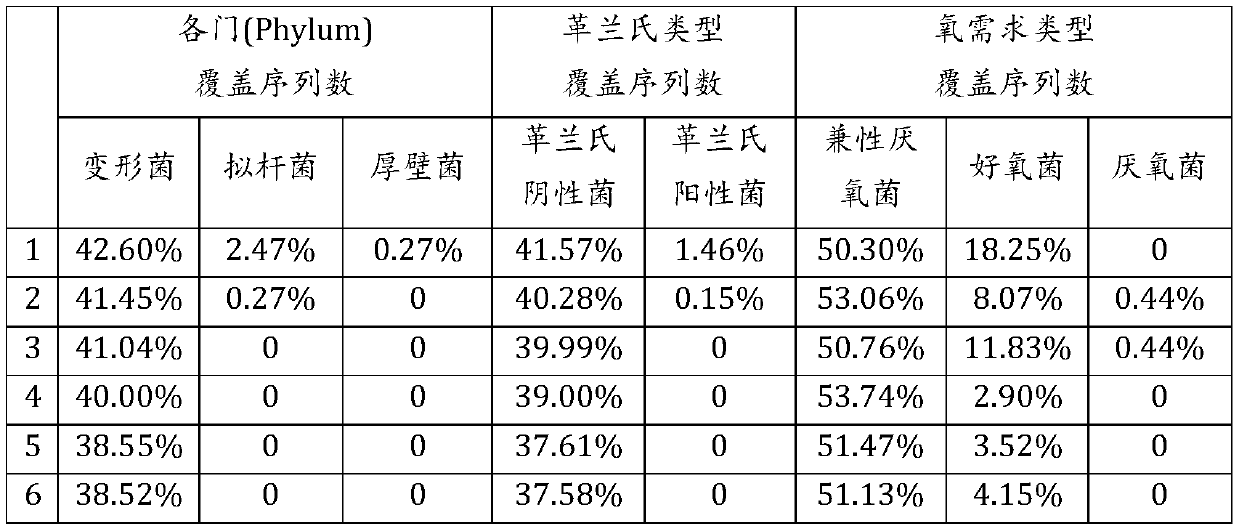
[0078] This experimental example is to use the nitrate reduction gene napA sequence database in the nitrogen cycle to design primers. The database includes 4562 napA gene sequences that are highly reliable and identified species (species). The database is mainly Proteobacteria, including More than 80% of the Proteobacteria, and the remaining 20% are composed of Firmicutes, Bacteroidetes and Chloroflexi; in addition, 58% of the sequences belong to facultative anaerobic groups in terms of oxygen demand types, 28% belonged to the aerobic group, 5% belonged to the anaerobic group; according to Gram classification, 82% of the sequences belonged to Gram-negative bacteria, and 15% belonged to Gram-positive bacteria.
[0079] Currently the most commonly used napA gene amplification primer pair is shown in SEQ ID NO: 1 and SEQ ID NO: 2. SEQ ID NO: 1 is V16cf-GCNCCNTGYMGNTTYTGYGG. In this sequence, N is A, T, C or G, and M is A or C, Y is C or T;
[0080] SEQ ID NO: 2 is: V17cr-RTGYT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com