In-vitro amplification detection method for novel coronavirus nucleic acid and test kit thereof
A viral nucleic acid, in vitro amplification technology, applied in microorganism-based methods, biochemical equipment and methods, and microbial determination/inspection, etc., can solve the problems of PCR product contamination, complicated operation, false positives, etc., and reduce PCR products. Contamination, high sensitivity, and the effect of improving specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
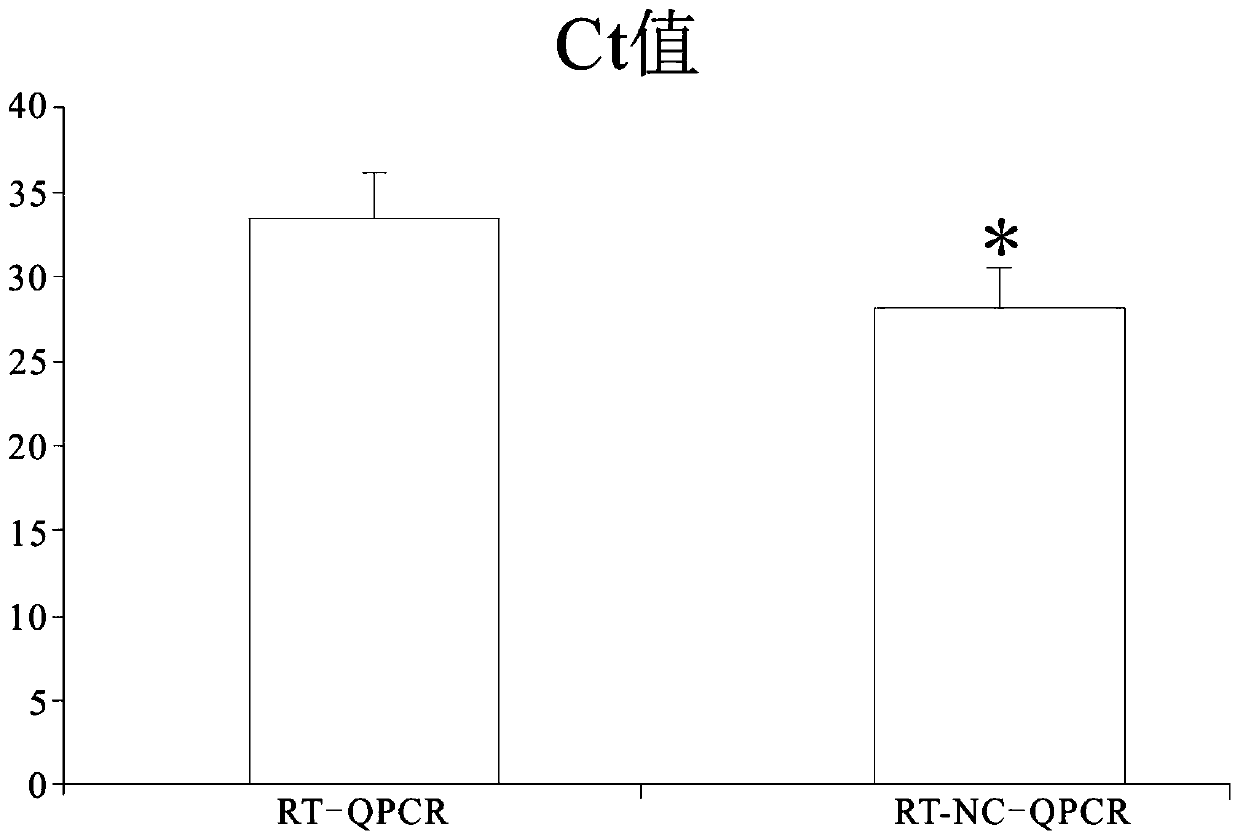
Image
Examples
Embodiment 1
[0022] 1. Materials
[0023] Human peripheral blood DNA was extracted by the conventional phenol-chloroform extraction method in our laboratory, and used as the template to be amplified.
[0024] 2. Primer design: Synthesize a pair of specific outer primers and a pair of special nested primers and complementary primers of the nested primers according to the sequence of the new coronavirus nucleic acid ORF1ab gene, and synthesize the following specific inner and outer primers:
[0025] Outer upstream primer P1: 5'-ACAACTTGTGCTAAT-3' (SEQ ID NO: 1);
[0026] Outer downstream primer P2: 5'-CATCAGCTGACTGAAG-3' (SEQ ID NO: 2);
[0027] Inner upstream primer P3: 5'-TTTTCCCTGTGGGTTTTACACTT-3' (SEQ ID NO: 3);
[0028] Inner downstream primer P4: 5'-TTTTTGGGTTCGCGGAGTTGAT-3' (SEQ ID NO: 4).
[0029] Complementary primer P3C of the inner upstream primer P3: 5'-AGTGTAAAACCCACAGG-3' (SEQ ID NO: 5);
[0030] Complementary primer of the inner downstream primer P4C: 5'-TCAACTCCGCGAACC-3'...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com