Multi-effector crispr based diagnostic systems
An effector protein, detection system technology, applied in applications, gene therapy, genetic engineering, etc., can solve problems such as limited operator availability, low sensitivity, and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0481]Example 1: Enhancing CAS13 activity with additional CRISPR-associated proteins
[0482] CRISPR effectors often interact with additional components to modulate activity, and applicants seek to exploit these interactions to increase the sensitivity and speed of SHERLOCK. Type VI-B CRISPR systems usually have interference regulatory proteins Csx27 and Csx28, and the co-expression of Csx28 has been shown to increase the interference activity of Cas13b protein in vivo, which means that they can increase the endonuclease activity of Cas13b in vitro. Since Csx28 was unstable in our hands, applicants purified the Csx28-Sumo fusion protein from three type VI-B systems ( Figure 5 A), and tested whether Csx28 supplementation increases the activity of Cas13a and Cas13b proteins. Applicants have found that the Csx28 protein either reduces the activity of Cas13 or increases target-independent (target-independent) cleavage ( Image 6 A-F).
Embodiment 2
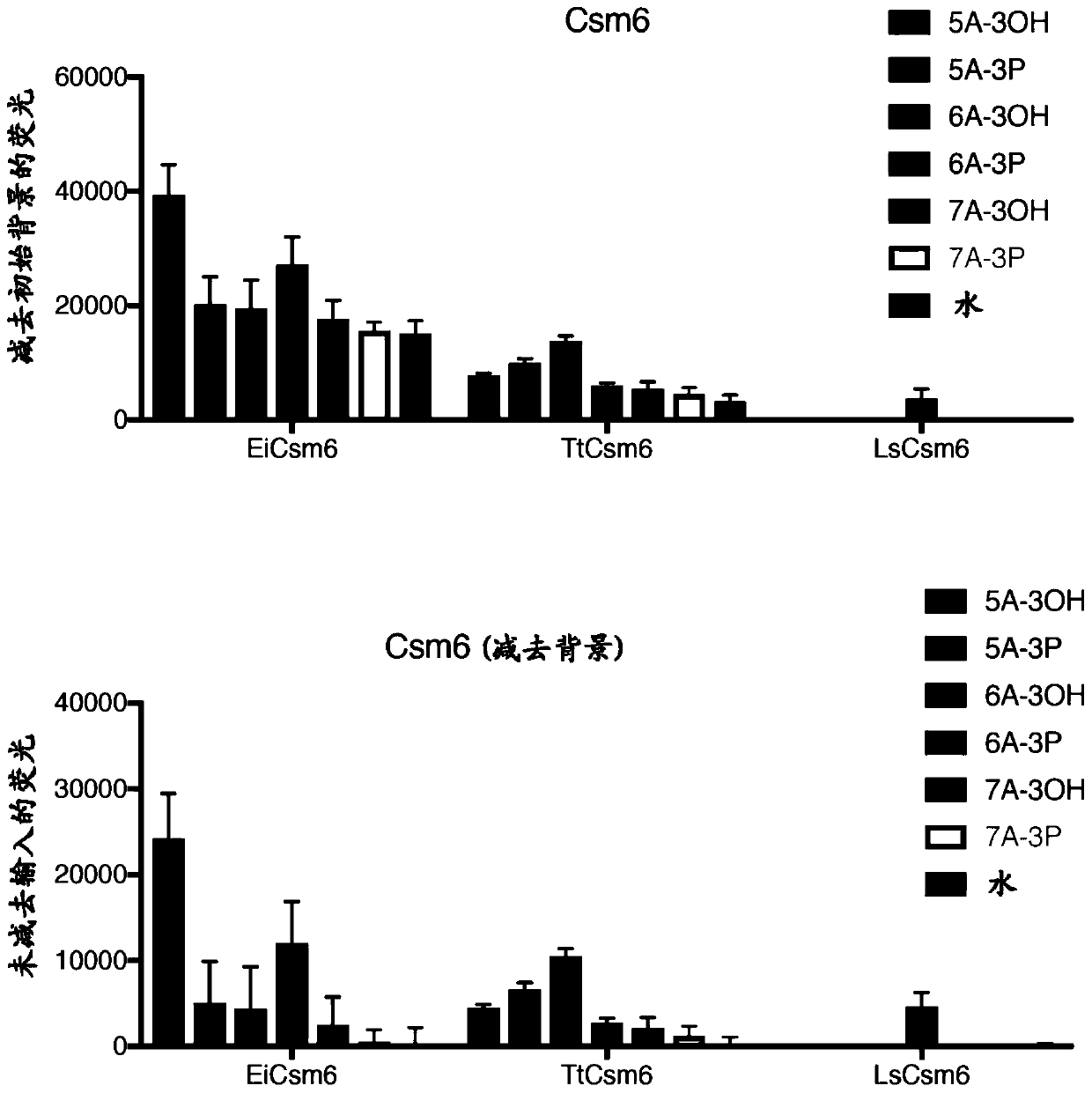
[0483] Example 2: Characterization of Csm6 Cleavage Activity
[0484] Interest was aroused by recent studies demonstrating nucleic acid-based allosteric activation of the CRISPR type III effector nuclease Csm6, and applicants wondered whether the endonucleic acid activity of Cas13 could generate a potent activator of Csm6 containing a 2,3-loop Linear RNA adenine phosphate ( Figure 7 C, 7D). Applicants therefore sought to characterize the RNA end chemistry of Cas13-derived cleavage products by performing a Cas13 in vitro cleavage assay on synthetic ssRNA 2 containing A or U homopolymer loops ( Figure 7 A). Post-cleavage fluorescent labeling of in vitro cleavage reactions confirmed that LwCas13a and PsmCas13b produced cleavage products with 5' hydroxylation and 2'3' cyclic phosphate RNA ends ( Figure 7 B). This result led us to express and purify both known and novel Csm6 orthologs for exploring the use of Csm6 together with Cas13 for positive feedback signal amplificatio...
Embodiment 3
[0485] Example 3: Positive feedback signal amplification using CRISPR-Csm6
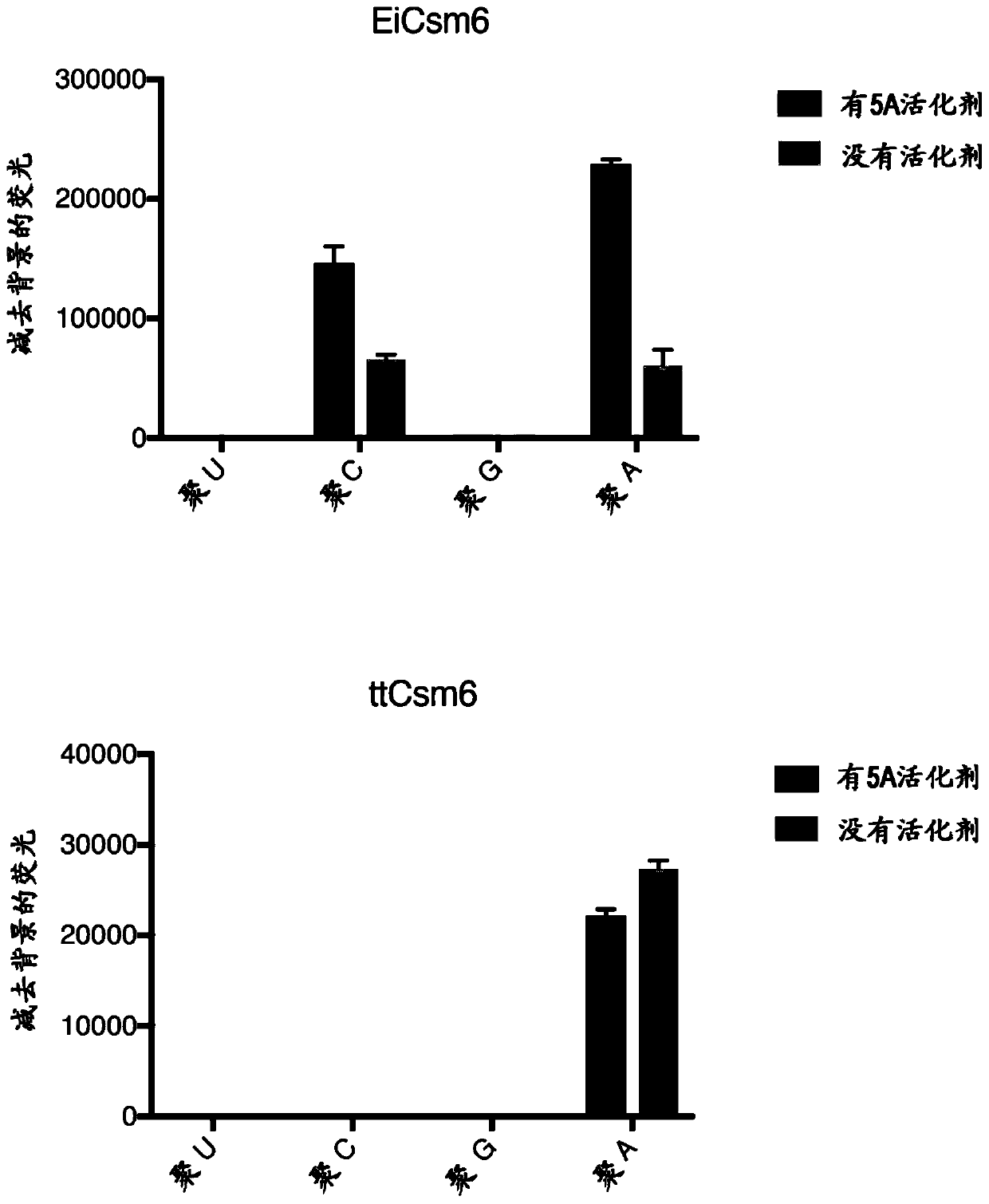
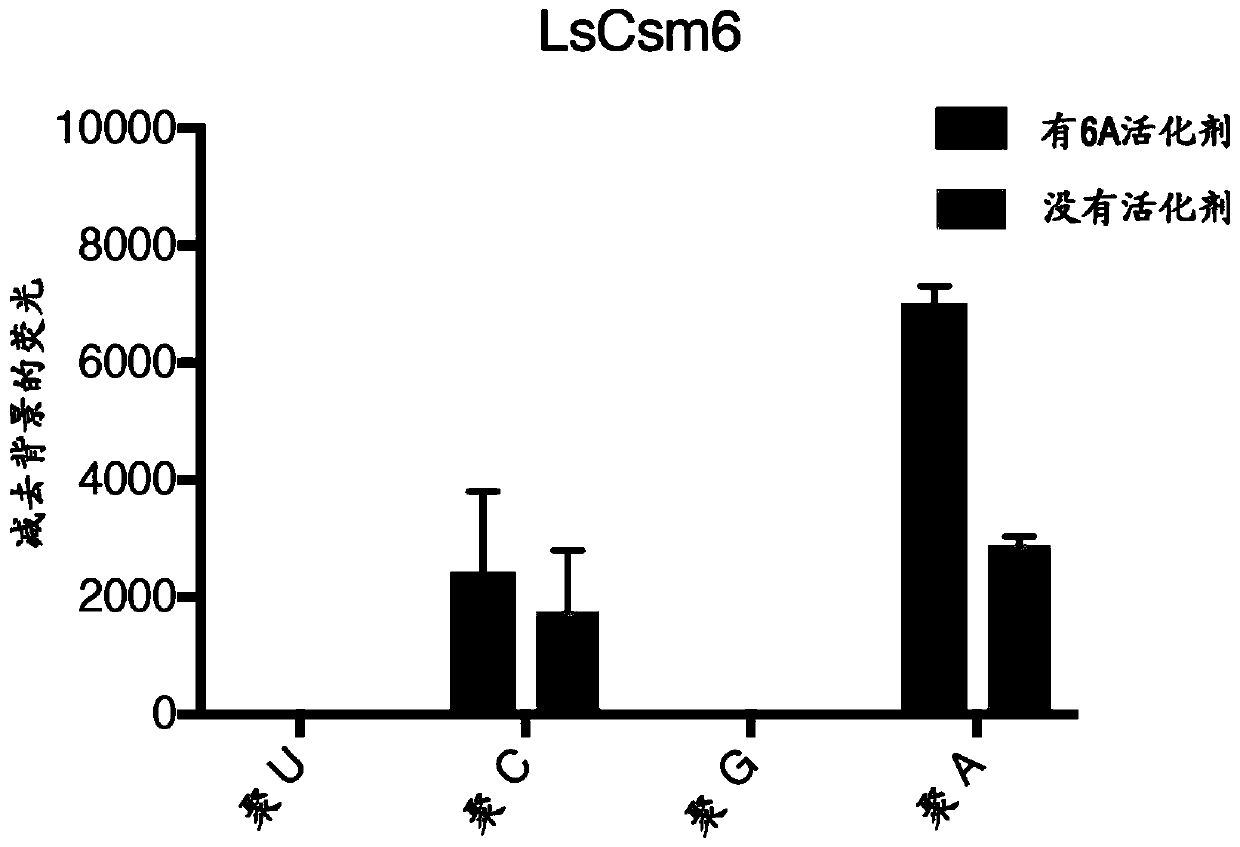
[0486] To couple Cas13 activity with Csm6 activation, Applicants designed RNA activators that would yield optimal Csm6 stimulation following Cas13 cleavage. Applicants incubated PsmCas13b with longer poly-A activators whose length is shortened by cleavage to generate short, cyclic phosphate-terminated activators. PsmCas13b digestion of the activator resulted in a modest increase in LsCsm6 activity ( Figure 14 A), possibly due to a series of sub-activators resulting from cleavage.
[0487] Applicants improved on this approach by designing RNA activators that contain two base properties (poly-A stretches with optimal activator length, followed by poly-U stretches that can be cleaved by U-targeting Cas13 enzymes). Applicants found that upon addition of the target, LwaCas13a was able to digest these activators and produce optimal activators against EiCsm6 and LsCsm6, and that this activation required t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com