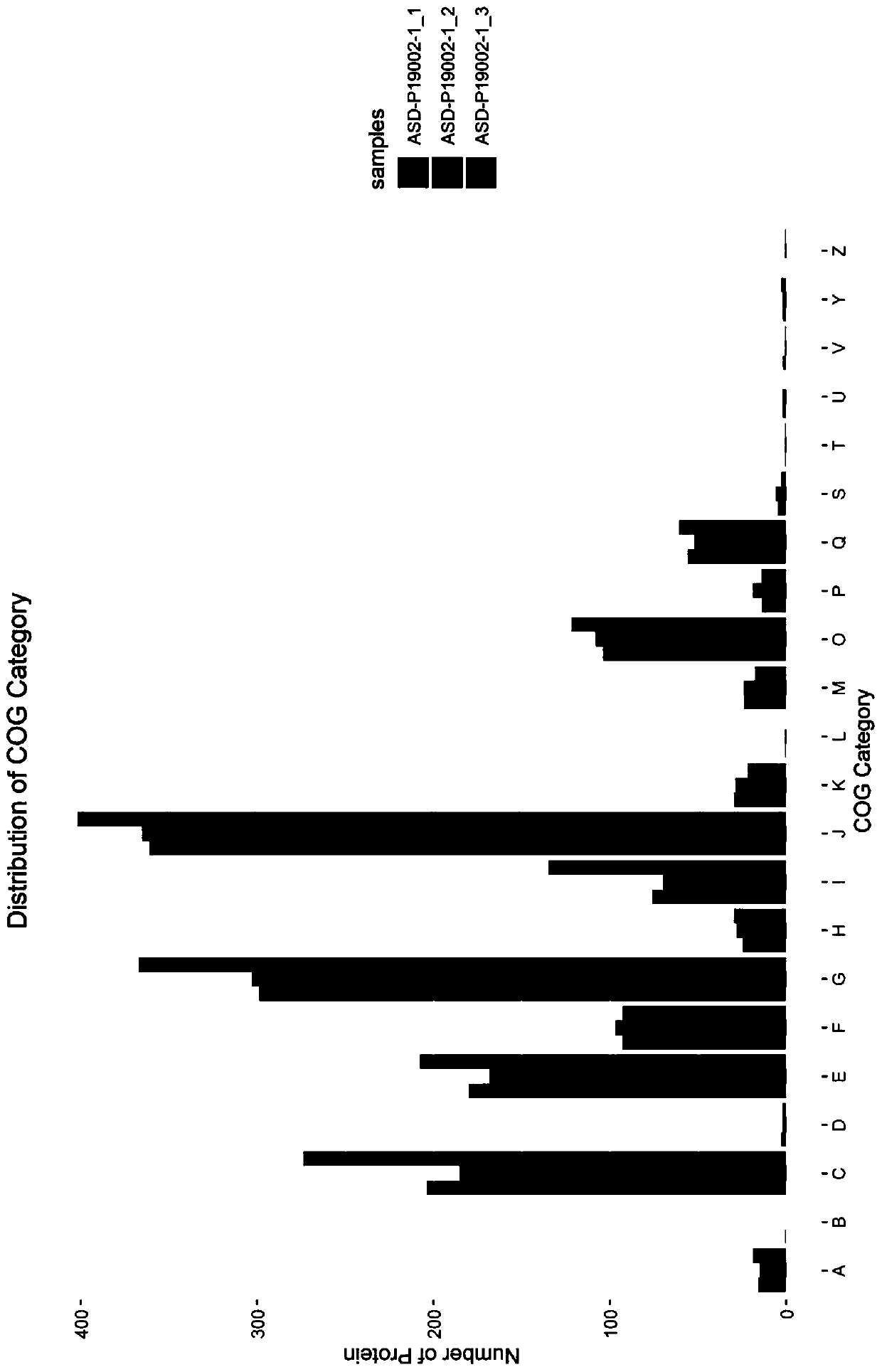
Metaproteome mass spectrometric data analysis method based on MetaPro-IQ method
A proteome and mass spectrometry data technology, applied in the field of bioinformatics, can solve problems such as high software and hardware requirements, long running time, redundant false positive results, etc., to achieve reduced volume, low hardware configuration requirements, and low processing time short-term effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0038] The technical solutions of the present invention will be described below in conjunction with the accompanying drawings and embodiments.
[0039] A method for analyzing metaproteome mass spectrometry data based on the MetaPro-IQ method, comprising the steps of:
[0040] Step S1 preliminarily streamlines the database: use X! for each piece of data to be tested separately. Tandem Alanine software searches and matches in the original database respectively, and the software parameters are set as follows:
[0041] parameter value fragment monoisotopic mass error 20 parent monoisotopic mass error plus 5 parent monoisotopic mass error minus 5 modification mass 57.022@C potential modification mass 16.0@M maximum missed cleavage sites 2 maximum valid expectation value 0
[0042] No result screening is performed during the search and matching process. After searching and matching, all matching results of each piece of data ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com