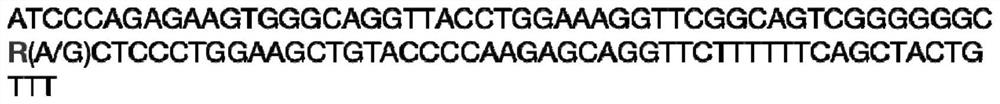A SNP Molecular Marker Associated with Feed Conversion Efficiency in Pigs
A feed conversion efficiency and molecular marker technology, applied in the field of swine molecular marker screening, can solve problems such as difficulty in improving feed efficiency-related traits, and inability to directly measure feed efficiency, and achieve high sensitivity and good specificity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1: Genotyping Detection
[0035] (1) Integrating 16 batches of SNP microarray data from 4005 pigs of four breeds including Large White pig, Landrace pig, Duroc pig and Pietrain pig collected by the laboratory;
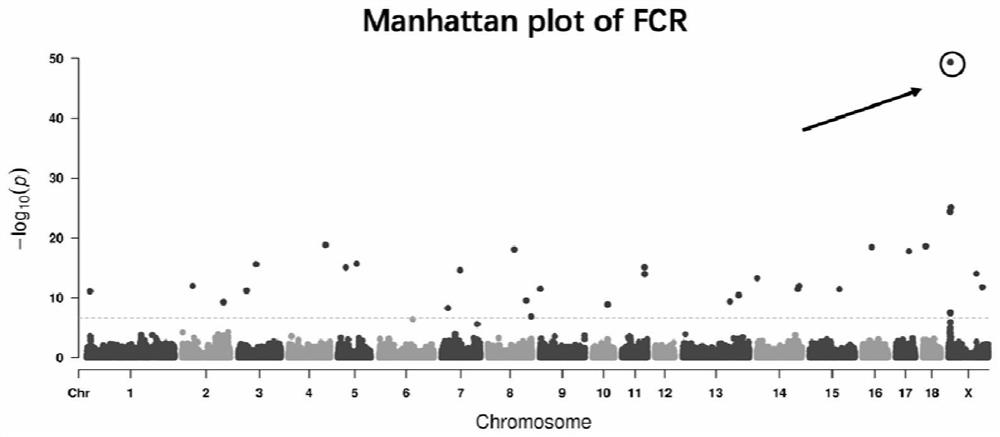
[0036] (2) Convert the raw data to Plink format (.ped / .map), and then use Plink software for preliminary quality control (--geno 0.1--maf 0.05--mind 0.1--chr 1-18,X). Use Beagle software to fill the data after the initial quality control, and then use Plink software for the second quality control (--geno 0.1--maf 0.05--mind 0.1), and finally there are 2013 individuals and 46505 SNPs for the whole Genome Association Analysis (GWAS).
Embodiment 2
[0037] Example 2: Application of MARC0045707 molecular marker typing method in pig feed conversion efficiency traits correlation analysis MARC0045707 molecular marker (see sequence listing SEQ ID NO: 1) and pig feed conversion efficiency traits correlation analysis:
[0038] The phenotypes used in the association analysis of genotypes and feed conversion efficiency traits mainly come from data collected in laboratories. Genome-wide association analysis (GWAS) was performed using the FarmCPU model in the rMVP package in the R statistical environment. The FarmCPU model uses a fixed effect model and a random effect model in a cycle. The specific models are as follows: (1) (2)y i = u i +e i , where y i Represents the trait observation value of the i-th individual, is the genotype of t possible association loci added to the model, b 1 , b 2 ,...,b t is the corresponding effect value of the possible association sites added to the model; S ij is the genotype of the jth gen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com