Micro RNA detection method based on one-step triggered branched DNA nanostructure
A nanostructure and tiny technology, applied in the direction of biochemical equipment and methods, microbial measurement/testing, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0028] The present invention will be described in further detail below in conjunction with the embodiments.
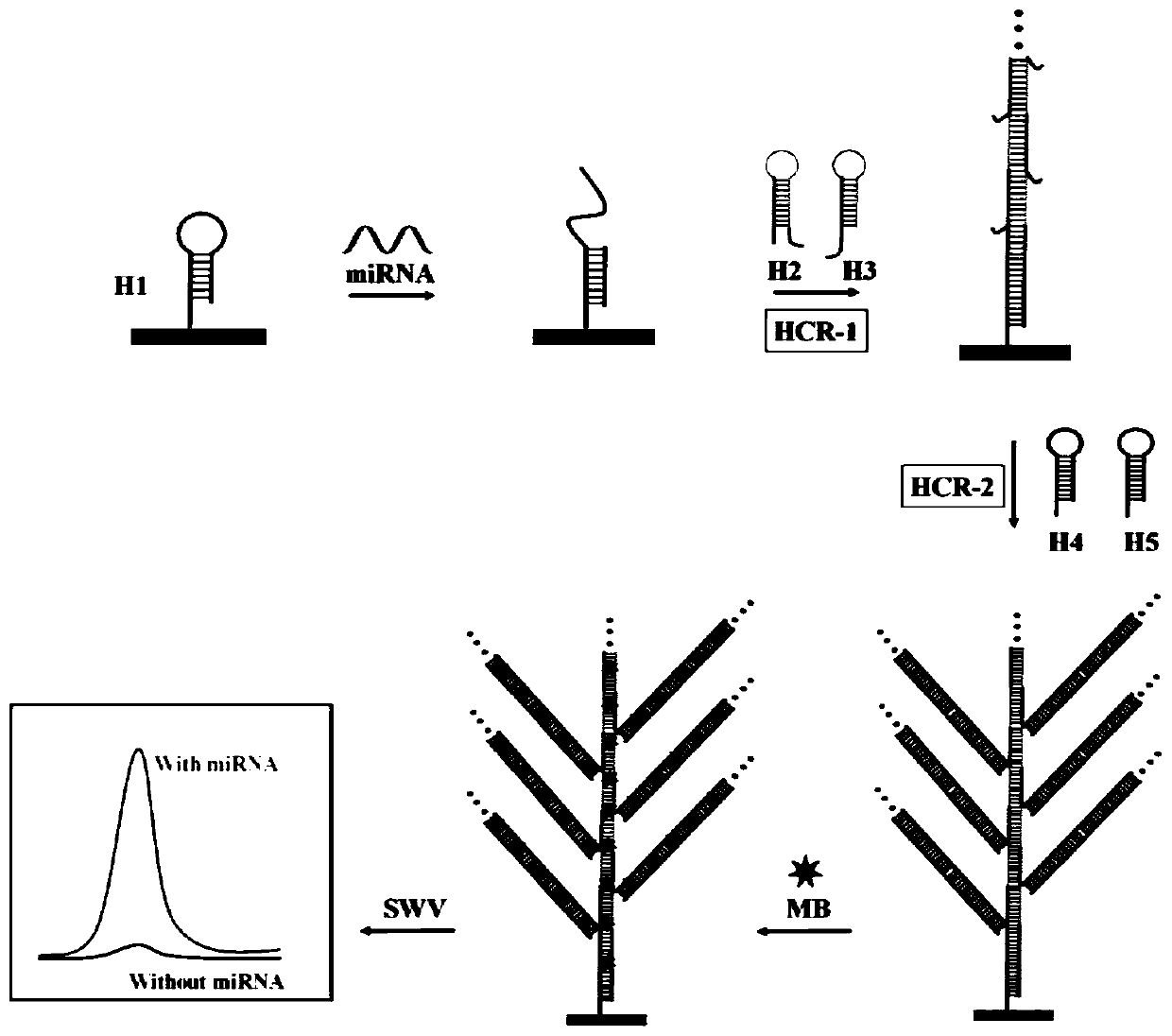
[0029] Such as figure 1 As shown, the principle of the present invention is: H1 is fixed on the surface of the gold electrode through the mercapto groups on the five initiator groups. MCH is then used to block the surface and reduce nonspecific adsorption, thereby increasing hybridization efficiency and accessibility of surface-bound hairpin DNA. Target microRNAs can hybridize to H1, thereby exposing the ends of H1. The end of the H1 is complementary to the toe and stem region of the H2, resulting in an opening of the H2 and exposing the end of the H2. The end of H2 can unfold H3 and release the end of H3. The release end of H3 is in the same order as the active part of H1. Thus, HCR-1 between H2 and H3 can be triggered to form a long nick of dsDNA flanked by some capture probes. Then, capture probes flanking the duplex can prime HCR-2 between two alternating hair...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com