Primers and kit for amplifying and detecting mutation of human COL1A1 and/or COL1A2 genes
A technology for sequencing primers and genes, which is used in DNA/RNA fragmentation, microbial determination/inspection, recombinant DNA technology, etc. It can solve the problems of low accuracy and specificity of determination results, incomplete detection results, and lack of amplification. , to save cost and manpower, reduce the number of PCR reactions, and simplify the amplification operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
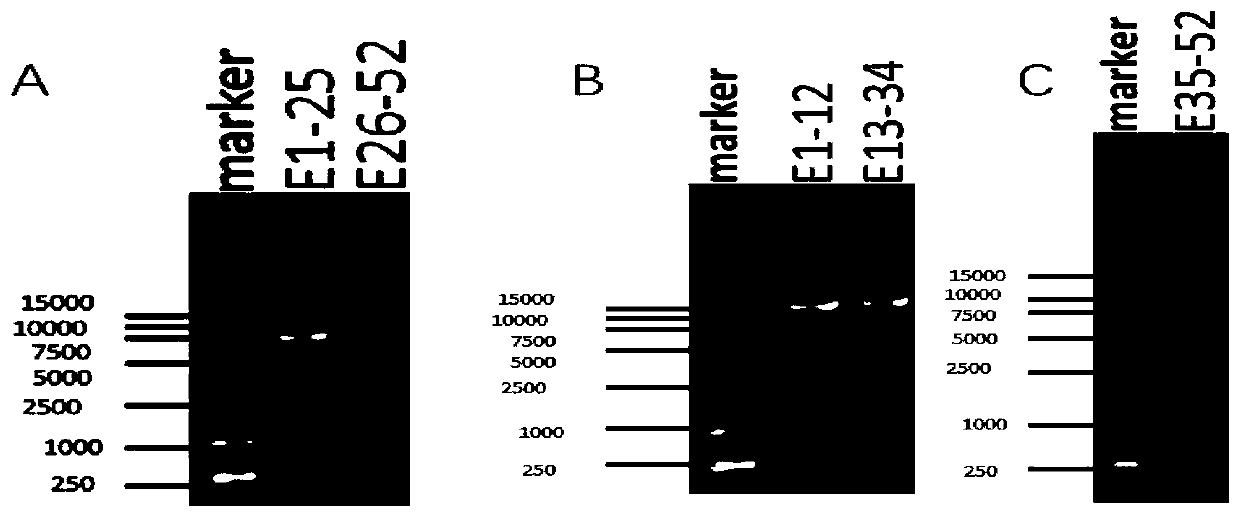
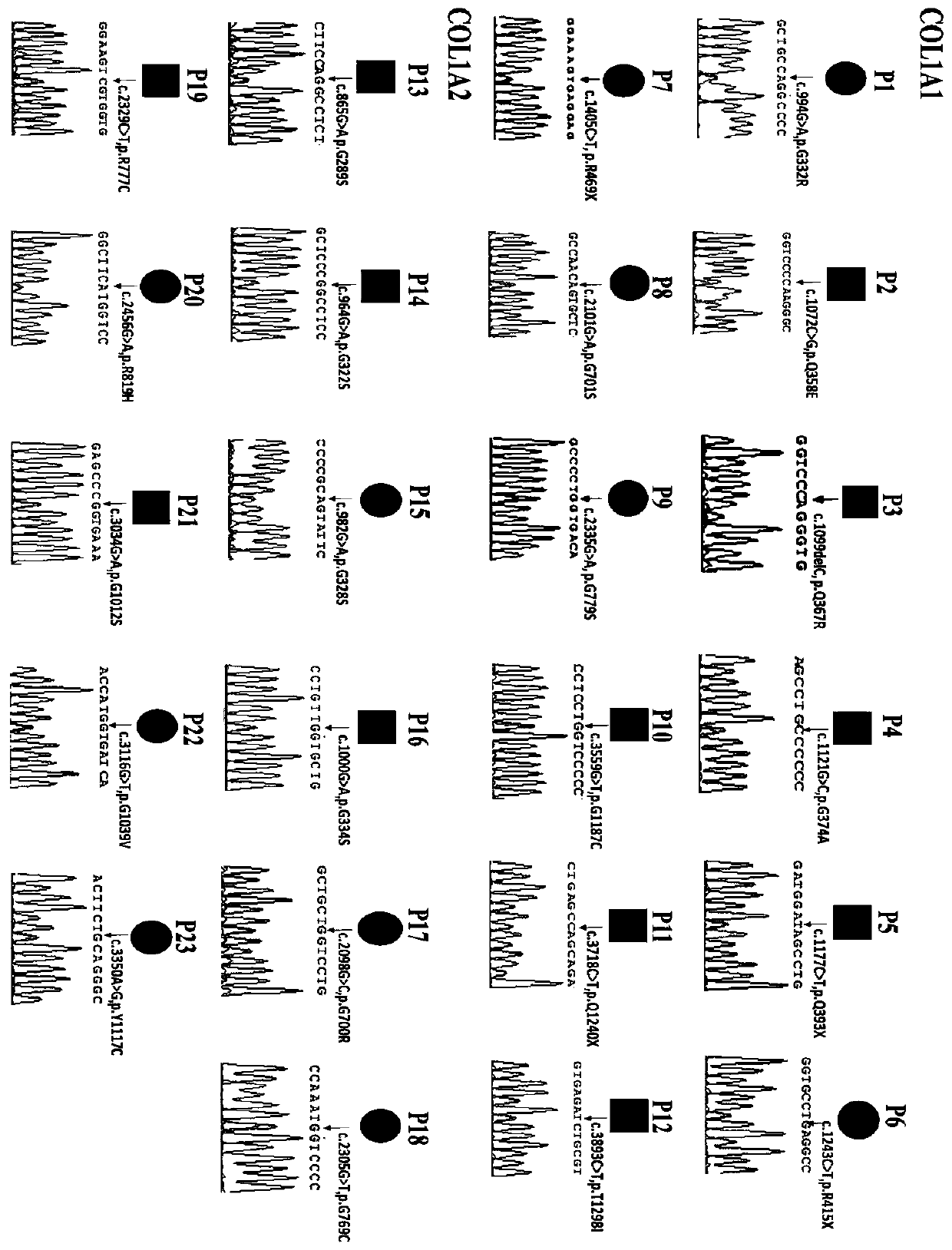
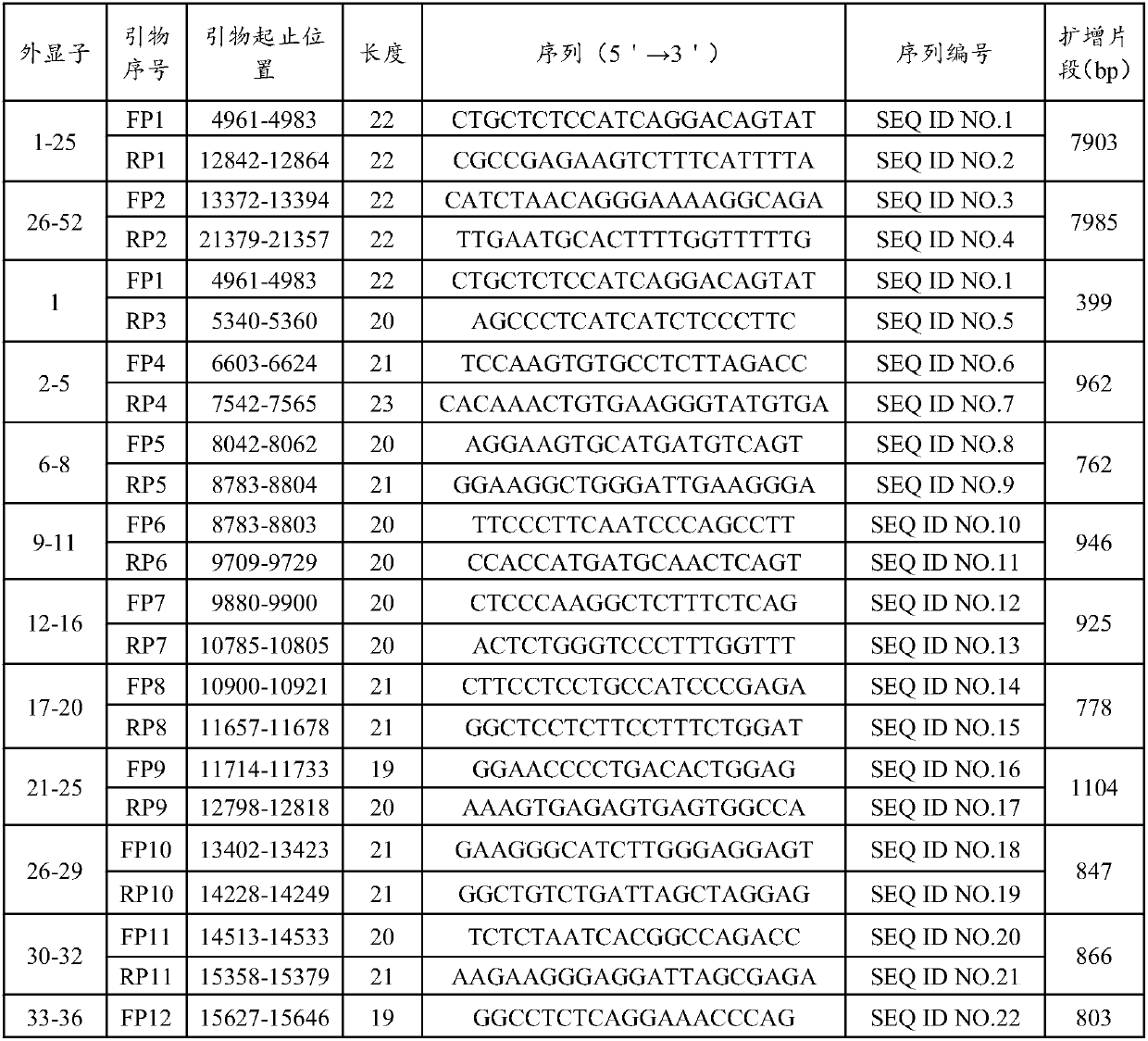
[0058] According to the NCBI GeneBank database COL1A1 (NG_007400.1) gene DNA reference sequence, design 2 pairs of long-segment PCR amplification primer pairs FP1 / RP1 and FP2 / RP2 to amplify the gene DNA sequence including all introns and exons of the gene . The exons covered by the two PCR products were 1-25 and 26-52, respectively, and the PCR amplification lengths were 7903bp and 7985bp, respectively. The PCR amplified fragments of each gene overlap each other by at least 150 bp to ensure that the subsequent upstream and downstream sequencing primers can detect each exon of the gene and its junction with the intron. For the exons included in the two long-fragment PCR amplifications, the upstream and downstream sequencing primers of the corresponding exons were designed respectively, and the sequencing primers were designed at the 60 bp upstream region at the junction of the exons and introns. Among them, there are 15 pairs of upstream and downstream sequencing sequences cor...
Embodiment 2
[0064] According to the DNA reference sequence of the COL1A2 (NG 007405.1) gene in the NCBI GeneBank database, three pairs of long-fragment PCR amplification primer pairs FP18 / RP18, FP19 / RP19, and FP20 / RP20 were designed to amplify all introns and exons of the gene. genetic DNA sequence. The exons covered by the three PCR products were 1-12, 13-34 and 35-52, respectively, and the PCR amplification lengths were 11549bp, 12218bp and 10502bp, respectively. The PCR amplified fragments of each gene overlap each other by at least 150 bp to ensure that the subsequent upstream and downstream sequencing primers can detect each exon of the gene and its junction with the intron. For the exons included in the three long-fragment PCR amplifications, the upstream and downstream sequencing primers of the corresponding exons were designed respectively, and the sequencing primers were designed at the 60 bp upstream region at the junction of the exons and introns. Among them, there are 31 pair...
Embodiment 3
[0071] A long fragment PCR amplification of human COL1A1 / COL1A2 gene DNA and a corresponding kit thereof, comprising:
[0072] 1) 2 pairs of primers FP1 / RP1 and FP2 / RP2 used for long fragment PCR amplification of COL1A1 gene in Example 1;
[0073] 2) 3 pairs of primers FP18 / RP18, FP19 / RP19 and FP20 / RP20 used in the long-fragment PCR amplification of the COL1A2 gene in Example 2;
[0074] 3) The PCR long fragment amplification reagent, including 10mM dNTP Mix, containing Mg 2+ 2×Phanta MaxBuffer, Phanta Max Super-Fidelity DNA Polymerase, RNase and DNase-free water; the working concentration of each component in the PCR amplification system is: 10000μM dNTP Mix, 2mM 2×Phanta Max Buffer, 1U / μlPhanta Max For Super-Fidelity DNA Polymerase, the upstream and downstream primers are both 5 μM, and the template concentration is 50-400 ng / μl.
[0075] A kit for detecting human COL1A1 / COL1A2 gene DNA mutation, comprising:
[0076] 1) The reagents for purifying long-segment PCR products...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com