Quantitative analysis method of Zygosaccharomyces bailii
A Bayer yeast, quantitative analysis technology, applied in the field of quantitative analysis of Bayer yeast, can solve the problems of difficult to distinguish, time-consuming, etc., and achieve the effect of stable primer amplification, high sensitivity, and good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1 Screening of the specific amplification site of Zygosaccharomyces bayeri
[0043] 1. Genome extraction of pure strains and screening of amplification sites
[0044] The 26S rRNA of each microorganism in Table 1 was extracted by conventional methods in the prior art, and all the extracted 26S rRNA sequences were compared to obtain a specific fragment of the 26S rRNA hypervariable region of Zygomyces bayerii with a length of 89 bp, such as SEQ ID NO: 3 shown.
[0045] 2. Specific Primer Synthesis
[0046] According to specific fragments, specific primers for Zygomyces bayeri 26S rRNA were synthesized, as shown in SEQ ID NO.1 and SEQ ID NO.2.
[0047] Bayer Zygomyces specific primer sequence (5'-3'):
[0048] SEQ ID NO.1
[0049] NLB-F:GAATCTCGCAGCT
[0050] SEQ ID NO.2
[0051] NLB-R: GTCCGCCACGAAGTGGT
[0052] 3. Select fluorescent quantitative PCR primers
[0053] The DNA of the main yeast strains of Moutai in Table 1 is divided into two groups, one gr...
Embodiment 2
[0068] The screening of the specific amplification primer of embodiment 2 Bayer yeast
[0069] Table 2. Real-time quantitative PCR primer sequences and screening results
[0070]
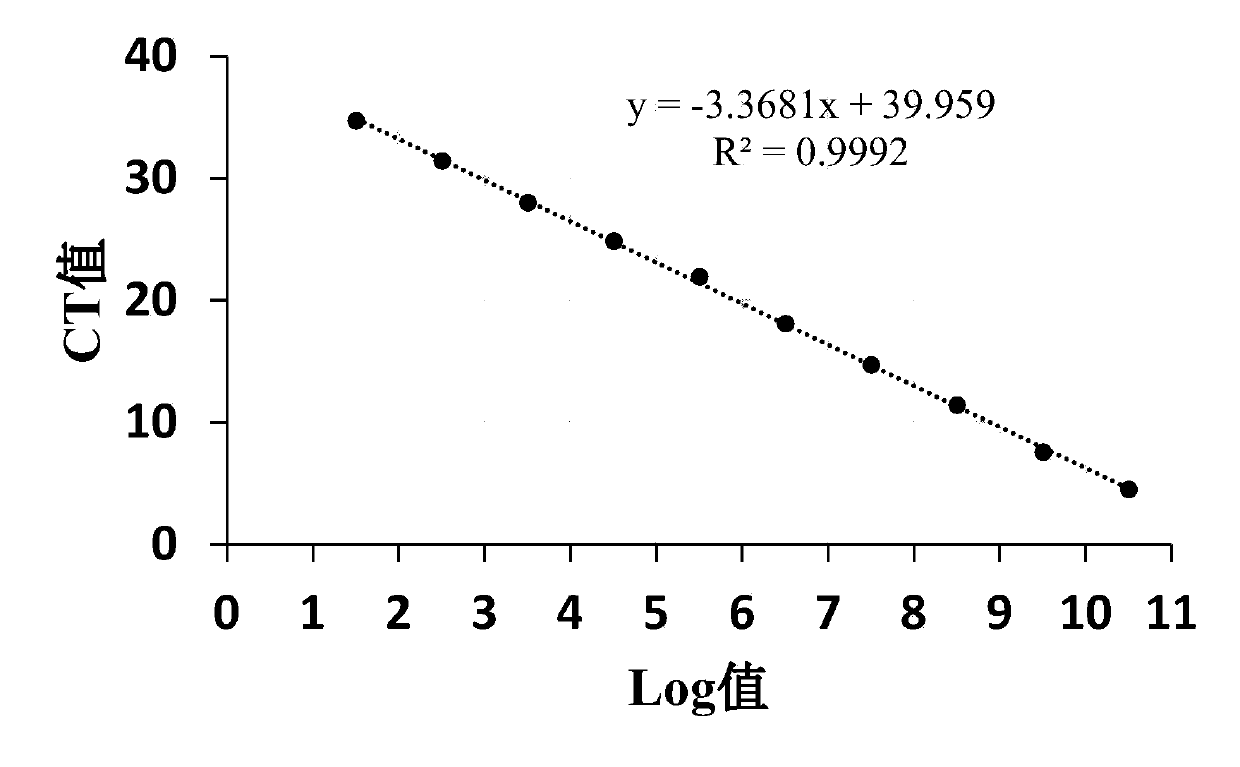
[0071] Since the real-time quantitative PCR is based on the relationship between the initial concentration of the template and the CT value, the optimization of the fluorescent quantitative PCR is very important to ensure that the sample has accurate and repeatable test results. The optimized real-time quantitative PCR has the following characteristics: (1) standard Curve R 2 >0.980; (2) High amplification efficiency (90–105%); (3) Consistent replicate reactions.
[0072] Amplification efficiency is the percentage of template amplified in each cycle, which is related to the slope of the standard curve. The calculation of the amplification efficiency of different primers in fluorescent quantitative PCR is as follows:
[0073] % efficiency = (10 -1 / 斜率 -1)×100%
[0074] Five pairs of primers wer...
Embodiment 3
[0076] Example 3 standard curve construction
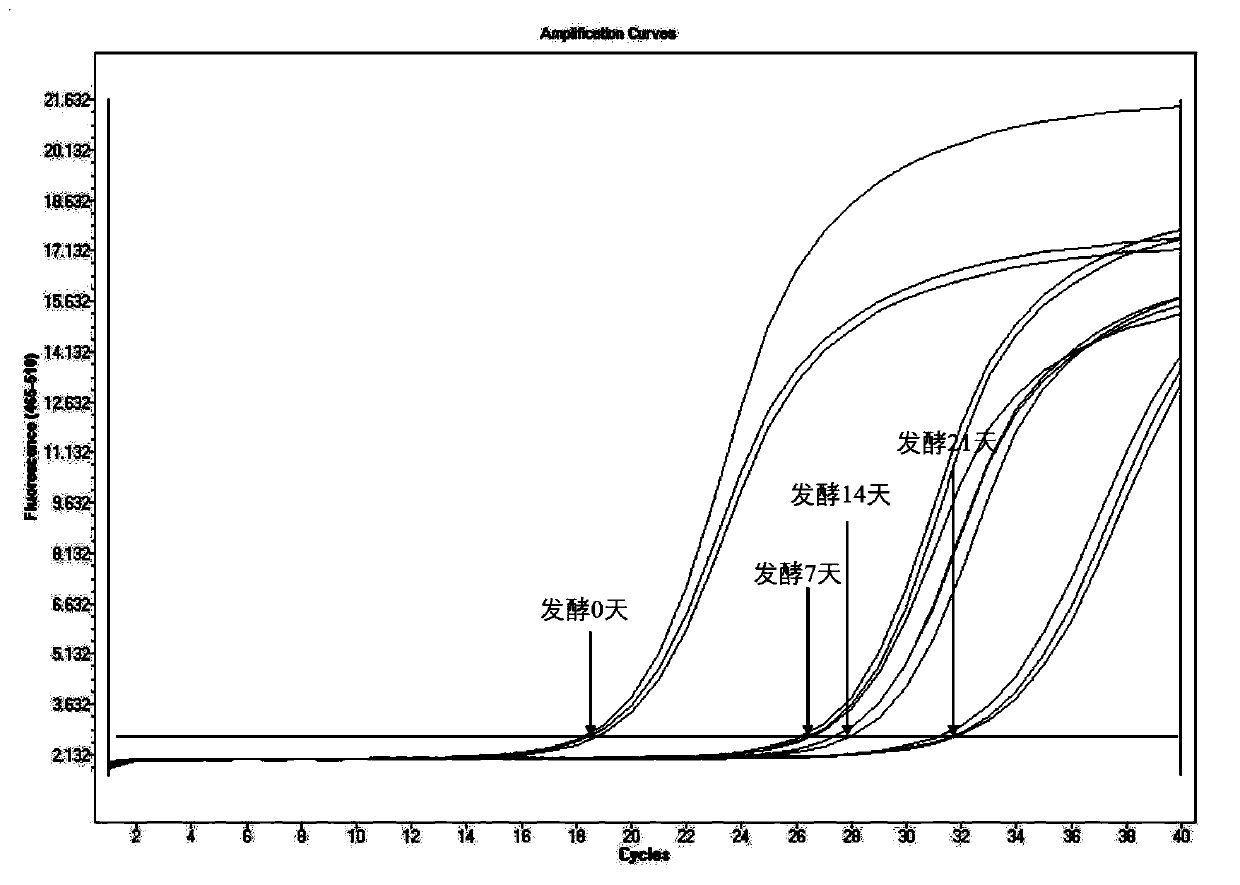
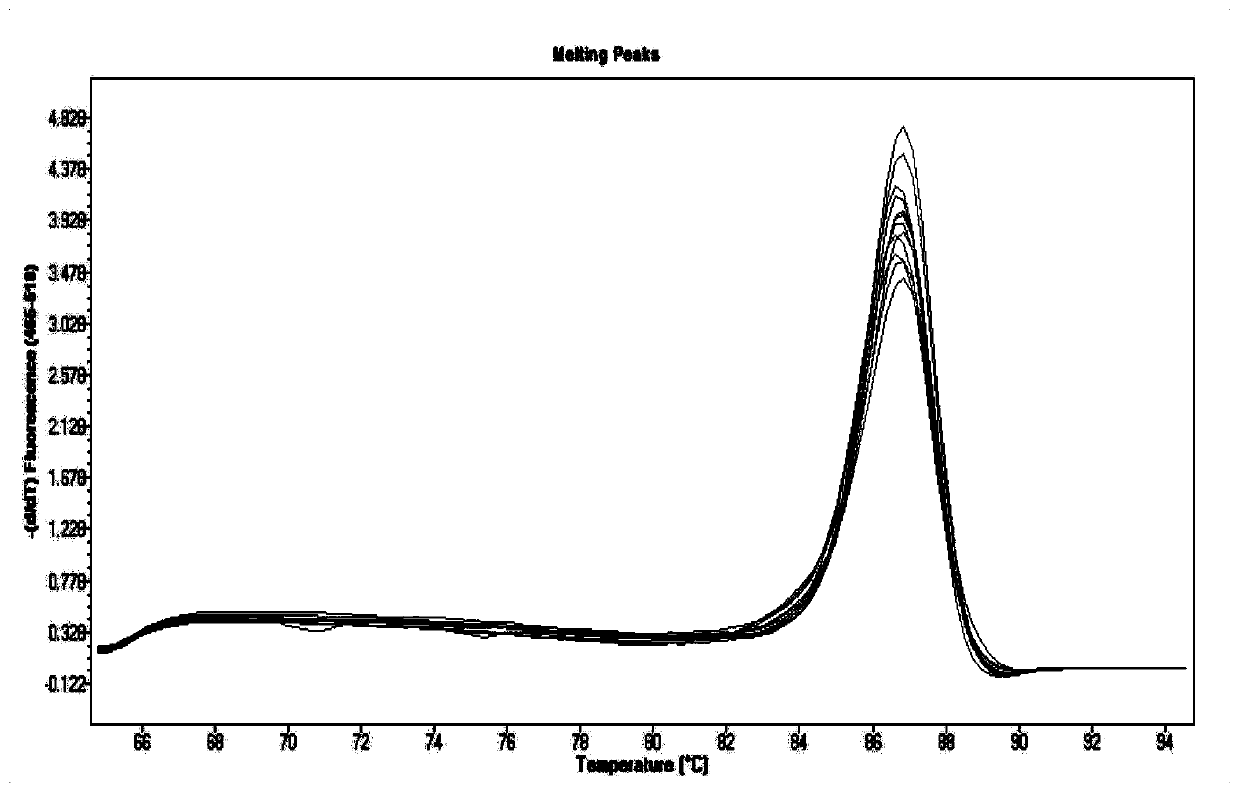
[0077] 1. Specific amplification of target fragments
[0078] The pure DNA of Zygomyces Bayer and the total DNA of fermented grains were used as templates, and NLB-F / NLB-R were used as primers for PCR amplification. PCR amplification system: total volume 25 μL, system including 2.5 μL Buffer, 2 μL dNTP, 0.2 μL Taq DNA polymerase, 1 μL 10 μmol / L primer (forward, reverse), 1 μL DNA template, with ddH 2 O to make up to 25 μL.
[0079] The PCR reaction program was as follows: 4 minutes of pre-denaturation at 95°C, followed by 30 cycles of 95°C for 10s; 50-52°C for 30s; 72°C for 10s, and finally 72°C for 7 minutes.
[0080] The amplified products were detected by 2% agarose gel electrophoresis and recovered by slicing the gel.
[0081] 2. Recombinant plasmid construction
[0082] The plasmid vector pUC57 and the target fragment were connected at a ratio of 1:1, and then the product was transformed into Mach1-T1 competent E. coli ce...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com