Combined genome for evaluating prognosis of clear cell renal cell carcinoma (ccRCC) and application of combined genome
A technology of renal clear cell carcinoma and genome, applied in a group of combined genomes and its application fields for evaluating the prognosis of renal clear cell carcinoma, can solve existing problems and other problems, and achieve the effect of improving survival rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
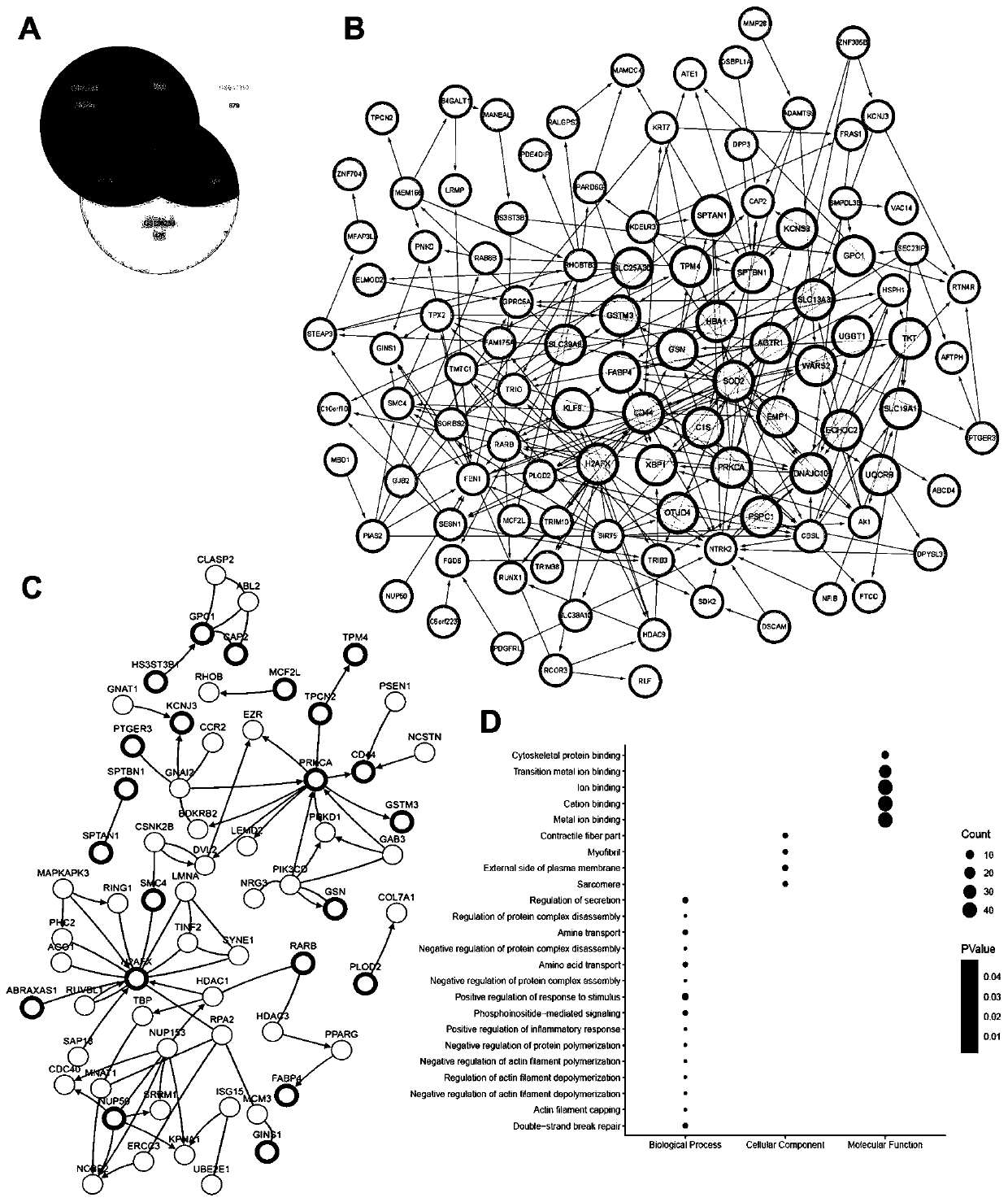
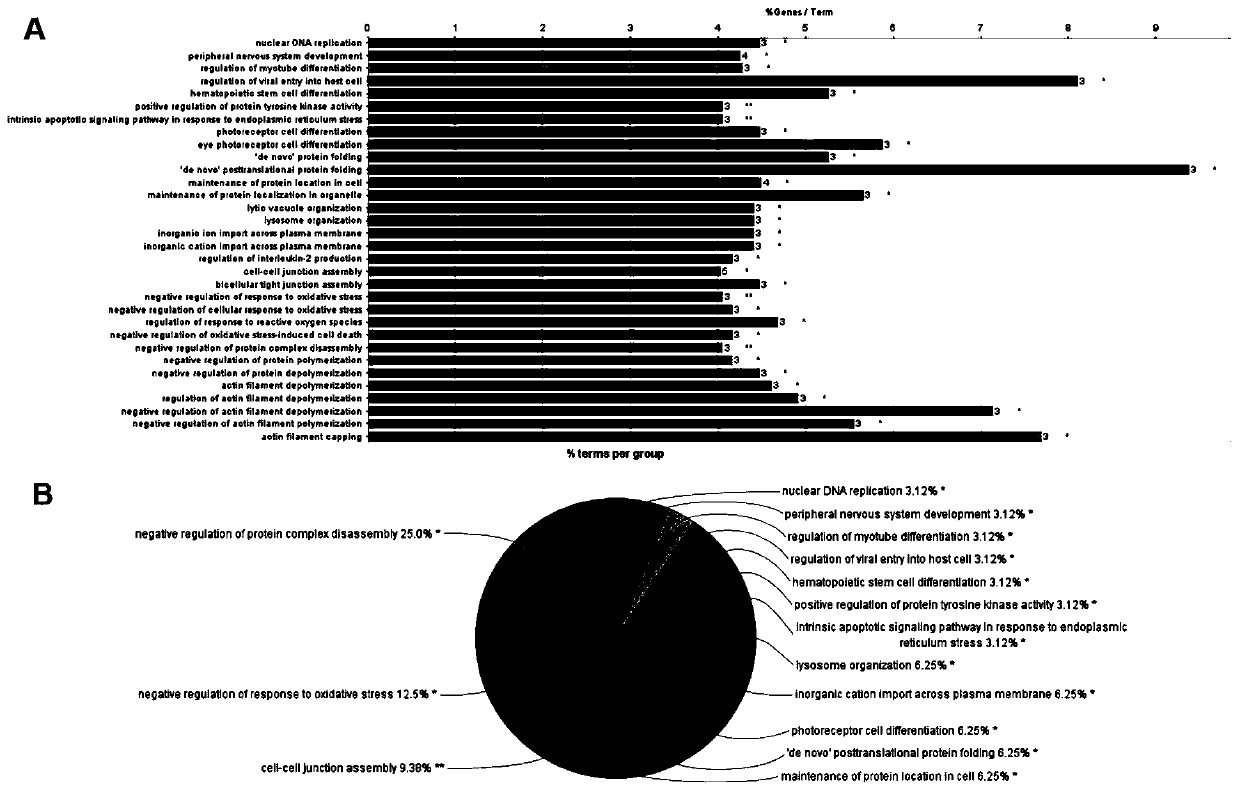
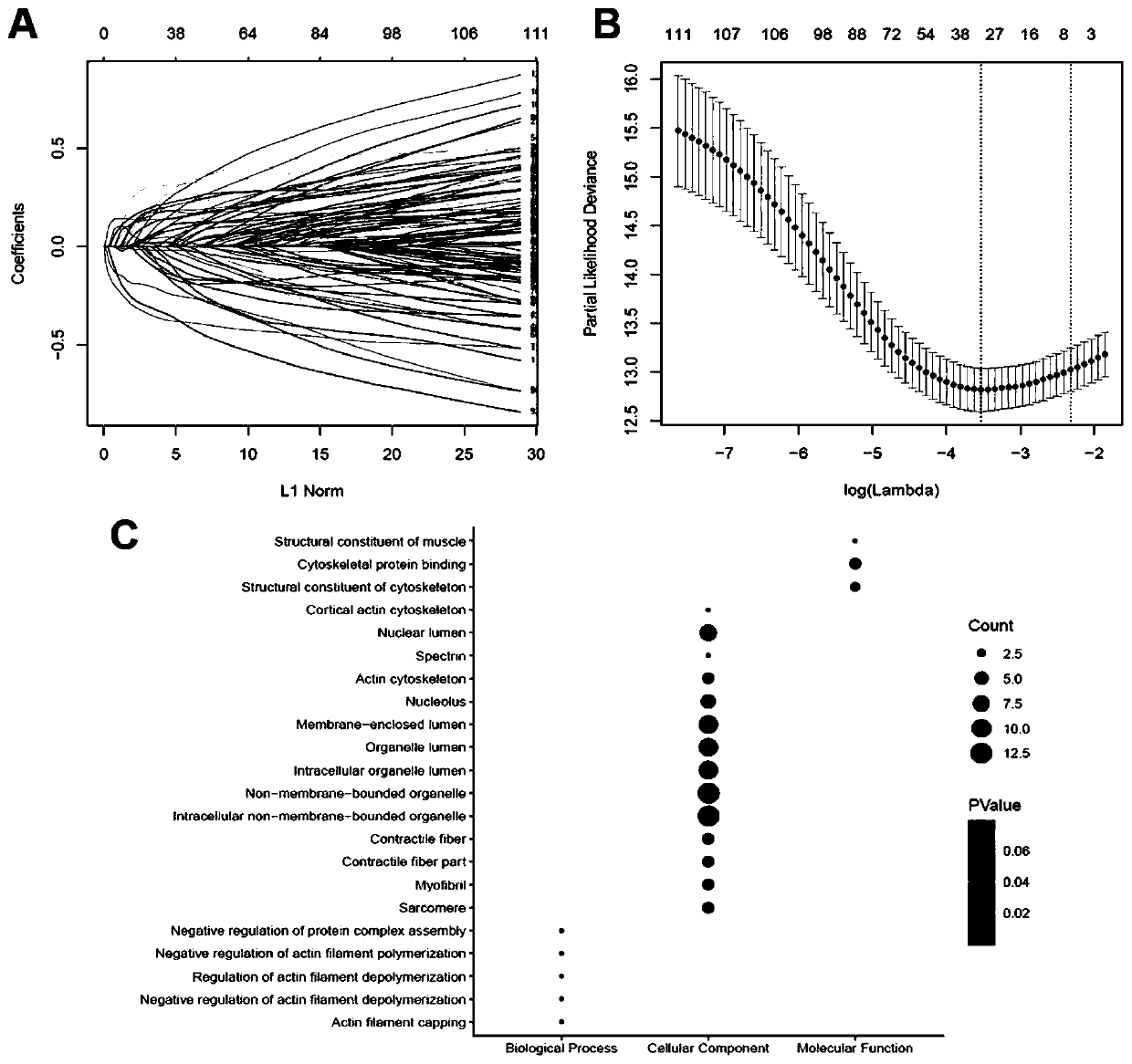
[0039] Example 1: Screening of ten marker genes
[0040] 1. Raw biological microarray data analysis
[0041] Raw transcriptional microarray data from metastatic or primary ccRCC samples were screened from the GEO (http: / / www.ncbi.nlm.nih.gov / geo) database. The corresponding genes converted into probes were converted into symbols with platform-processed annotation information. Three chip datasets GSE22541, GSE47352 and GSE85258 were downloaded from the same GEO platform using Affymetrix Human Genome U133 Plus 2.0 arrays (24 primary and 44 metastatic tumor samples in GSE22541, 4 primary tumor samples in GSE47352 and 4 metastatic tumor samples, and 14 primary and 14 metastatic tumor samples in GSE85258).
[0042] 2. Normalization and clarification of DEG
[0043] Preprocessing and normalization of raw biological data is the first step in processing DNA microarrays. This process removes bias from microarray data to ensure its uniformity and integrity. Probe data were subseque...
Embodiment 2
[0063] Example 2 External Verification
[0064] From June 2009 to September 2017, 380 surgical specimens of ccRCC patients were selected from the Urology Department of Shanghai Cancer Center, Fudan University. Tissue samples including ccRCC and normal tissues were collected during surgery and are available from the FUSCC tissue bank.
[0065] 1. Real-time quantitative PCR (RT-qPCR) analysis
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com