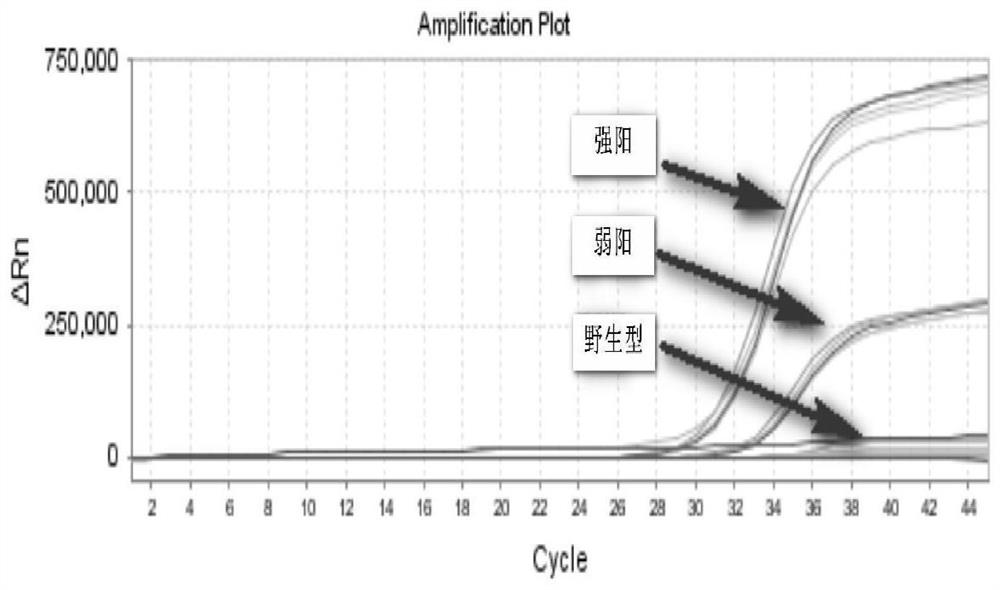
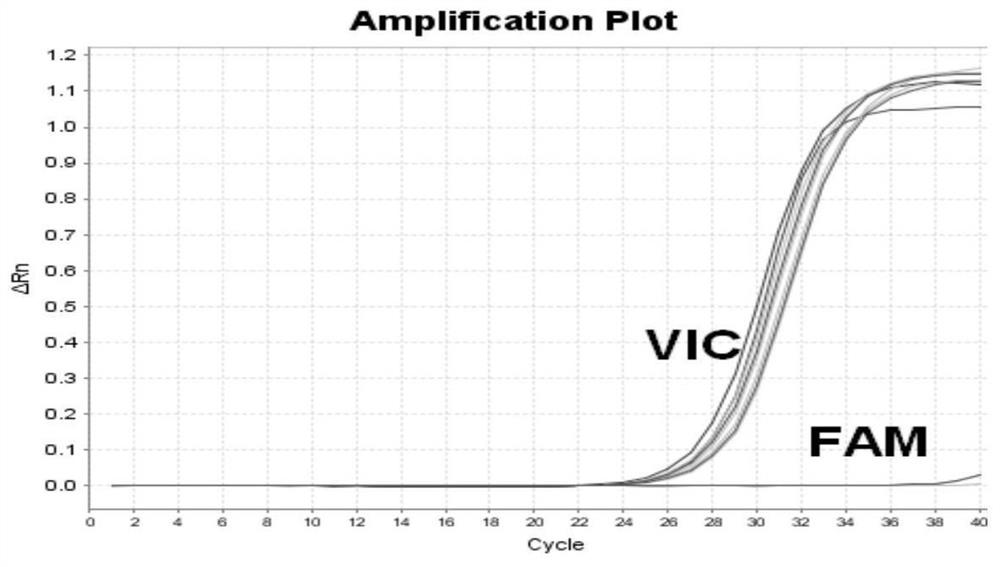
Primers for detecting mutation of human B-raf gene V600E, primer probe composition and kit
A primer-probe and kit technology, applied in recombinant DNA technology, microbial assay/inspection, biochemical equipment and methods, etc., can solve the problems of poor wild-type background tolerance, low amplification efficiency, and high detection cost , to achieve the effect of short detection time, high amplification efficiency and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
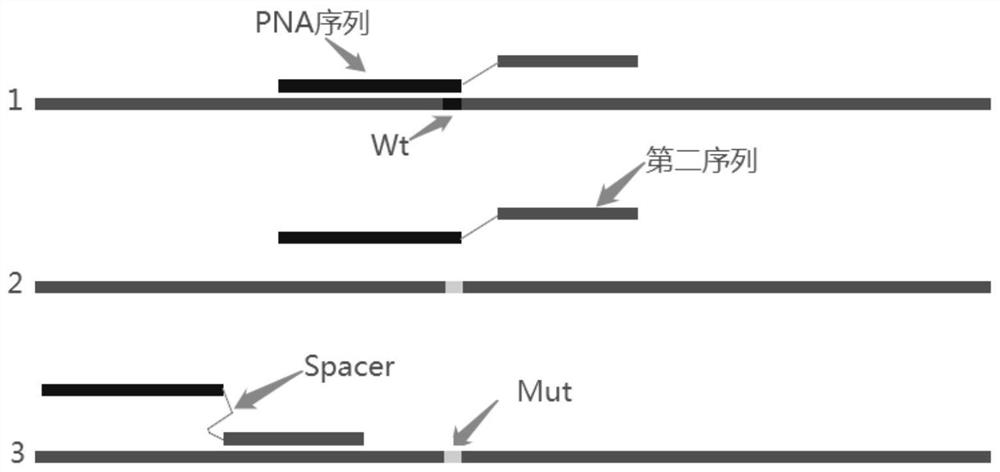
[0066] This embodiment provides a primer for detecting the V600E mutation region of the B-raf gene, including a forward primer and a reverse primer. The forward primer includes three parts arranged in the following order along the 5' end to the 3' end:
[0067] 1) The first sequence, the peptide nucleic acid PNA sequence used to identify mutation hotspots and hybridize with the wild-type template, which completely matches the wild-type template sequence and has one base mismatch with the mutant template sequence, the nucleotide sequence is as follows Shown in SEQ ID NO.2;
[0068] SEQ ID NO.2: 5'-ATCGAGATTTCACT-3' (Tm: 65°C)
[0069] 2) Spacer, which connects the 3' end of the first sequence and the 5' end of the second sequence, is C6;
[0070] 3) The second sequence is combined with the sequence upstream of the mutation site, 3-6 bp of its 3' end sequence overlaps with the 5' end of the first sequence, and the nucleotide sequence is shown in SEQ ID NO.4;
[0071] SEQ ID NO...
Embodiment 2
[0076] This embodiment provides a primer for detecting the V600E mutation region of the B-raf gene, including a forward primer and a reverse primer. The forward primer includes three parts arranged in the following order along the 5' end to the 3' end:
[0077] 1) The first sequence, the peptide nucleic acid PNA sequence used to identify mutation hotspots and hybridize with the wild-type template, which completely matches the wild-type template sequence and has one base mismatch with the mutant template sequence, the nucleotide sequence is as follows Shown in SEQ ID NO.1;
[0078] SEQ ID NO.1: 5'-TCGAGATTTCACTGTAGCTA-3' (Tm: 76°C)
[0079] 2) Spacer, connecting the 3' end of the first sequence and the 5' end of the second sequence, which is C3;
[0080] 3) The second sequence, combined with the sequence upstream of the mutation site, 3-6 bp of its 3' end sequence overlaps with the 5' end of the first sequence, and the nucleotide sequence is shown in SEQ ID NO.5;
[0081] SEQ...
Embodiment 3
[0086] This embodiment provides a primer for detecting the V600E mutation region of the B-raf gene, including a forward primer and a reverse primer. The forward primer includes three parts arranged in the following order along the 5' end to the 3' end:
[0087] 1) The first sequence, the peptide nucleic acid PNA sequence used to identify mutation hotspots and hybridize with the wild-type template, which completely matches the wild-type template sequence and has one base mismatch with the mutant template sequence, the nucleotide sequence is as follows Shown in SEQ ID NO.3;
[0088] SEQ ID NO.3: 5'-CATCGAGATTTCACT-3'
[0089] 2) Spacer, which connects the 3' end of the first sequence and the 5' end of the second sequence, is C18;
[0090] 3) The second sequence, combined with the sequence upstream of the mutation site, 3-6 bp of its 3' end sequence overlaps with the 5' end of the first sequence, and the nucleotide sequence is shown in SEQ ID NO.6;
[0091] SEQ ID NO.6: 5'-CTGA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com