Inducible DNA methylation editing system based on CRISPR/dCas9
An editing system and methylation technology, applied in the biological field, can solve the problems of off-target and inability to realize time-space specific DNA methylation editing, etc., and achieve the effect of wide application range and flexible use method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
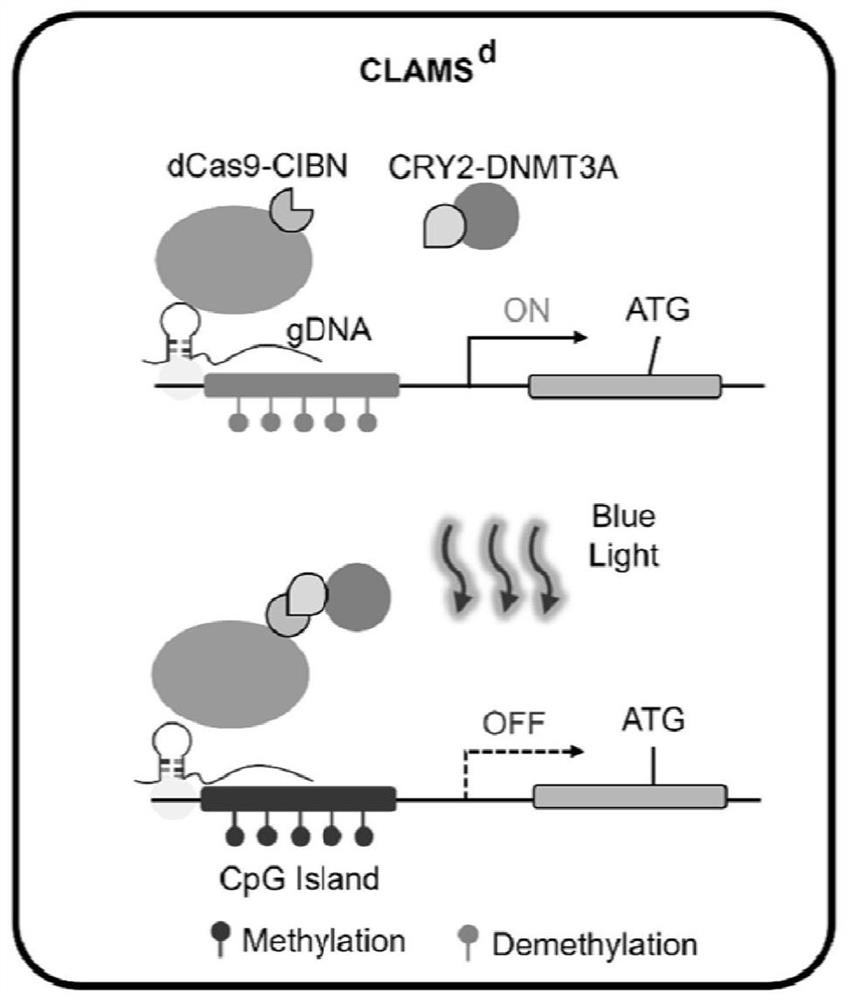
[0035] Embodiment 1: light-controlled DNA methylation writing system (CLAMS d ) construction
[0036] Such as figure 1 Shown, CLAMS dConsisting of a three-plasmid system, SpCas9D10A and H840A are mutated to lose endonuclease activity but retain DNA targeting activity, so it is called deadCas9 (dCas9), and the SV40NLS-FLAG-dCas9-CIBN-NLS fusion protein is constructed using the whole gene synthesis method As the anchor element, CRY2PHR was fused to the DNA methyltransferase DNMT3A as the effector element on the other hand. The Cas9 gRNA clone is downstream of the human U6 promoter and expressed by U6. Under dark conditions, gRNA guides the dCas9-CIBN protein to the designated position of the genome, but the DNA methylation effect element is free in the nucleus; under blue light irradiation, CIBN recruits CRY2PHR and carries the activation element to the designated position to edit DNA methylation .
Embodiment 2
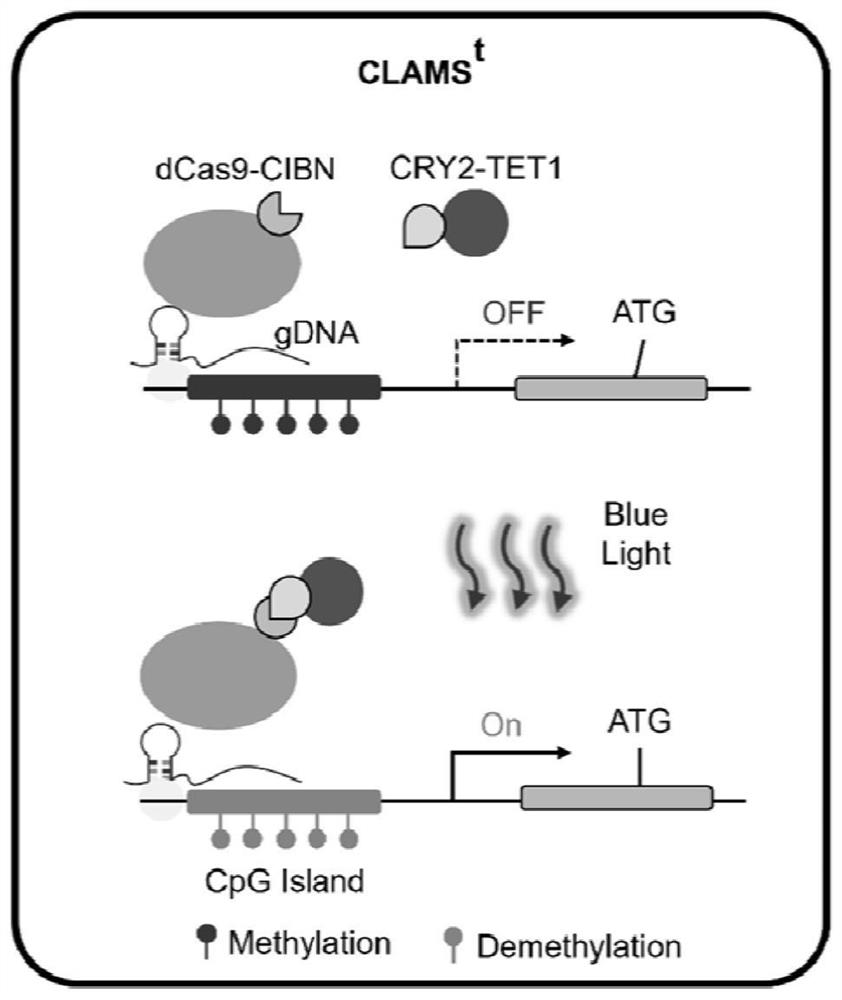
[0037] Embodiment 2: light-controlled DNA demethylation system (CLAMS t ) construction
[0038] CLAMS t with CLAMS d The composition is similar, the principle is as figure 2 Shown, CLAMS d It also consists of a three-plasmid system, using the whole gene synthesis method to construct the SV40NLS-FLAG-dCas9-CIBN-NLS fusion protein as the anchor element, and on the other hand, the fusion of CRY2PHR and the demethylase TET1 as the effector element. The Cas9 gRNA clone is downstream of the human U6 promoter and expressed by U6. Under dark conditions, gRNA guides the dCas9-CIBN protein to the designated position of the genome, but the DNA methylation effect element is free in the nucleus; under blue light irradiation, CIBN recruits CRY2PHR and carries the activation element to the designated position, modifying DNA demethylation the process of.
Embodiment 3
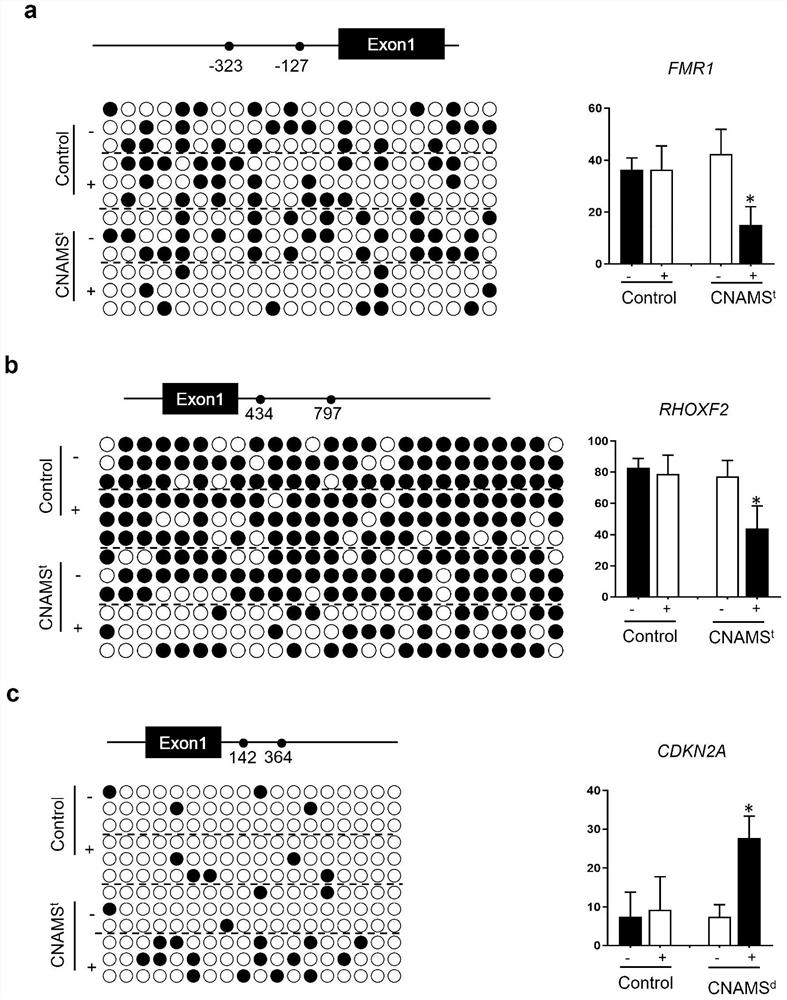
[0039] Example 3: Verification of the CLAMS DNA methylation editing system
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com