Proteome mass spectrometric data processing method and device
A technology of proteome and mass spectrometry data, applied in the field of bioinformatics, can solve the problems of unreliable results, result differences, normalization, removal of batch effect differences, calculation and selection methods without consistent standards, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
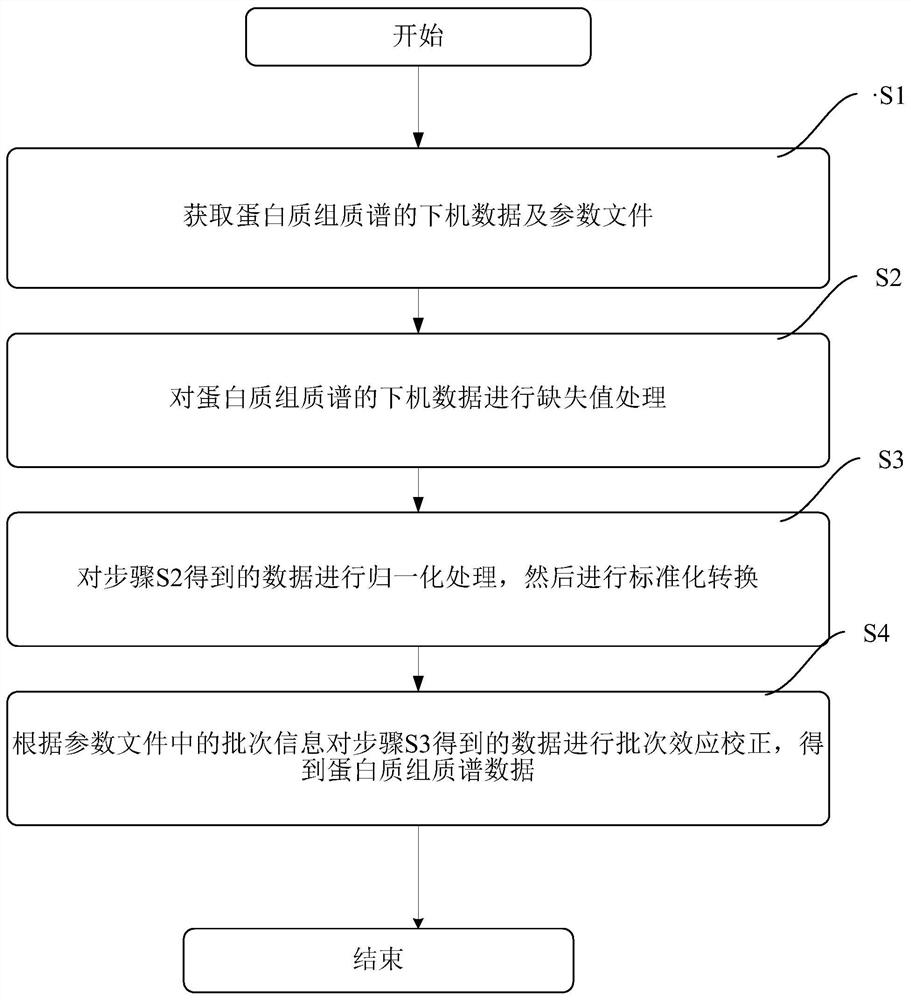
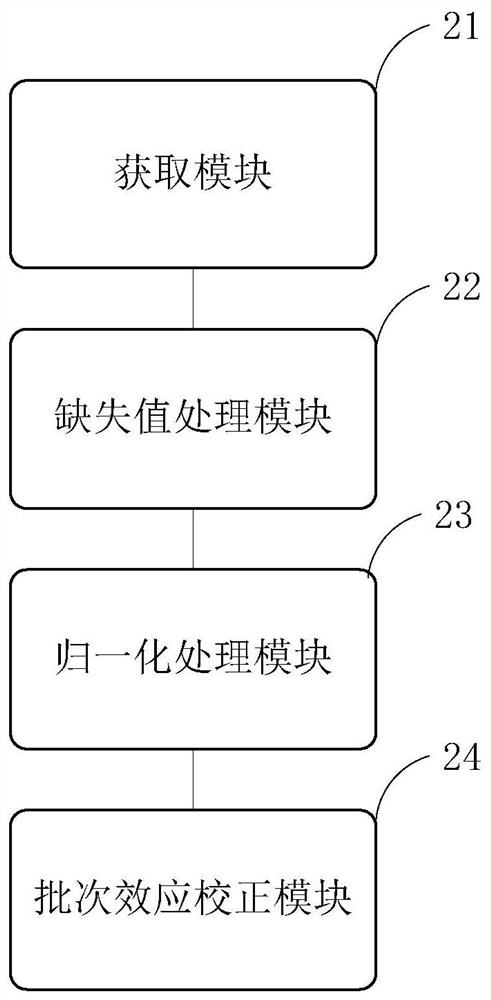
[0136] 1. Data preparation
[0137] The input files accepted by the present invention are off-machine data of proteome mass spectrometry (the name of the protein must be in the official standard gene symbol format) and parameter files.
[0138] 1.1 The proteome mass spectrometry data are as follows (the example is the proteome data of Escherichia coli in different culture media):
[0139]
[0140] 1.2 Run parameter input (all characters are English characters):
[0141] project_name = "Proteome_test";
[0142] project_dir=" / home / test / Proteome";
[0143] KEGG_enrichment = "eco";
[0144] GO_enrichment="org.EcSakai.eg.db";
[0145] norm_method="loess";
[0146] runDifferential = TRUE;
[0147] enrichment_qval = 0.05;
[0148] DEG_logFC = 1;
[0149] DEG_qval = 0.05;
[0150] …
[0151] 2. Data preprocessing
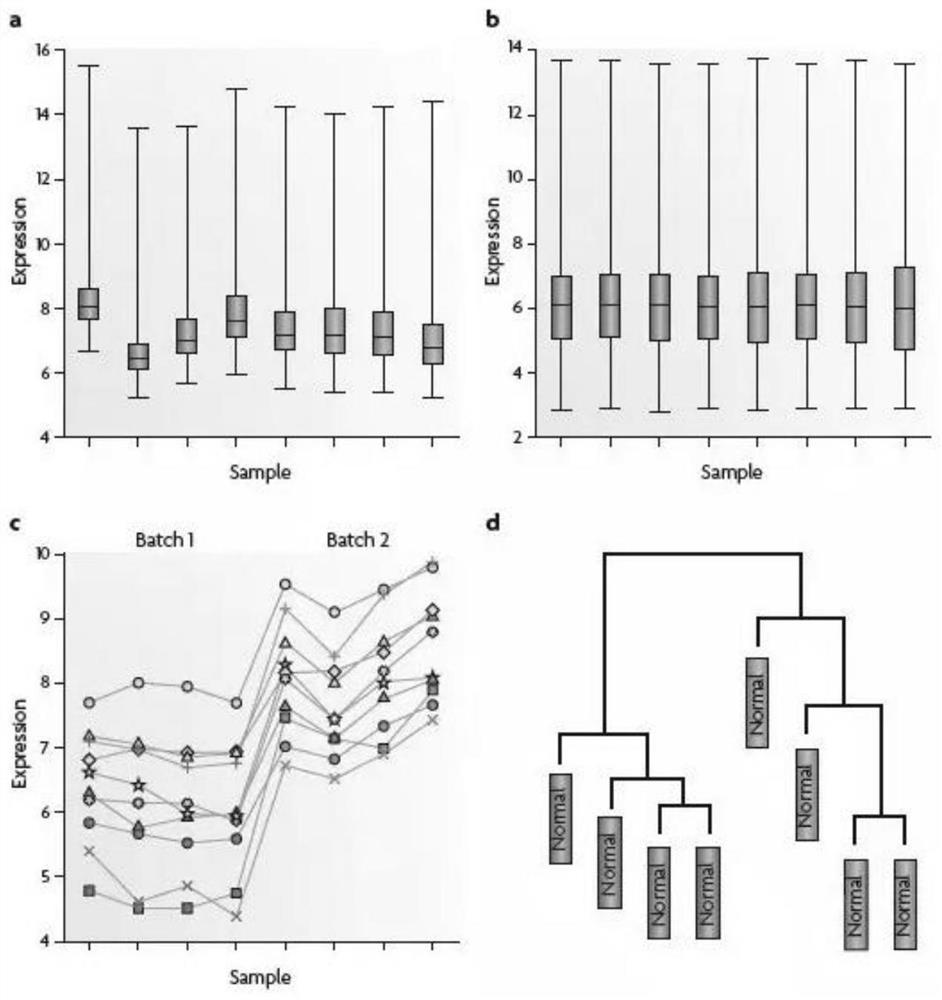
[0152] Handle missing values, perform the first overall quality analysis on the original data, and then perform LOESS normalization processing on the proteomi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com